Abstract
RNAs 1 and 2 of the tripartite genome of alfalfa mosaic virus encode the replicase proteins P1 and P2, respectively, whereas RNA 3 encodes the movement protein and coat protein. Transient expression of wild-type (wt) and mutant viral RNAs and proteins by agroinfiltration of plant leaves was used to study cis- and trans-acting functions of the helicase-like domain in P1 and the polymerase-like domain in P2. Three mutations in conserved motifs of the helicase-like domain of P1 affected one or more steps leading to synthesis of minus-strand RNAs 1, 2, and 3. In leaves containing transiently expressed P1 and P2, replication of wt but not mutant RNA 1 was observed. Apparently, the transiently expressed P1 could not complement the defect in replication of the RNA 1 mutant. Moreover, the transiently expressed wt replicase supported replication of RNA 2, but this replication was blocked in trans by coexpression of mutant RNA 1. However, expression of mutant RNA 1 did not interfere with the replication of RNA 3 by the wt replicase. Similarly, a mutation in the GDD motif encoded by RNA 2 could not be complemented in trans and affected the replication of RNA 1 by a wt replicase, while replication of RNA 3 remained unaffected. In competition assays, the transient wt replicase preferentially replicated RNA 3 over RNAs 1 and 2. The results indicate that one or more functions of P1 and P2 act in cis and point to the existence of a mechanism that coordinates the replication of RNAs 1 and 2.
Viruses with a plus-strand RNA genome that is larger than 5.8 kb generally encode a protein or protein domain which has homologies to known helicases (18). Helicases catalyze DNA and/or RNA duplex unwinding, and to that end bind and hydrolyze ATP as an energy source (reviewed in reference 4). Based on sequence comparisons, helicase-like proteins have been classified into four superfamilies (SF), of which SF 1 and 2 are most abundant (4, 12, 18). The best-studied viral helicase is that of hepatitis C virus (HCV), family Flaviviridae, which is a DEA(D/H) box RNA helicase of SF 2 (references 22 and 34 and references therein). Mutation of two residues in the helicase domain of HCV nonstructural protein 3 (NS3) inhibited virus replication in vivo (19). Also, it has been shown that the NS3 helicases of bovine viral diarrhea virus (BVDV) and dengue virus type 2, both members of the family Flaviviridae, are required for virus and/or RNA replication (15, 16, 23). Moreover, mutation of the helicase function of BVDV NS3 was found to inhibit negative-strand RNA synthesis (16).
Alfalfa mosaic virus (AMV) is a positive-strand RNA virus with a tripartite genome (for a review, see reference 3). It belongs to the family Bromoviridae. RNAs 1 and 2 encode the replicase proteins P1 and P2, respectively. Of these, P1 contains homologies to known methyltransferase domains in its N-terminal half (28, 41) (Fig. 1A) and an SF 1 helicase-like domain in its C-terminal half (12, 18) (Fig. 1A). P2 is the viral RNA-dependent RNA polymerase (RdRp) protein (26). AMV RNA 3 codes for the movement protein (P3) and the coat protein (CP), which is translated from subgenomic RNA 4. Viruses of the alphavirus-like superfamily of viruses, to which AMV belongs, generally encode helicase (or helicase-like) proteins of SF 1 (18). Duplex-unwinding activities of several viral SF 1 helicases have been characterized in the past few years (2, 11, 13, 31, 32). However, little is known about the roles of these helicases in viral life cycles. It has been shown that the NTPase activities or NTP-binding motifs of the helicase domains of the 126-kDa protein of tobacco mosaic virus (TMV), the nsP2 helicase of Semliki Forest virus, p150 of turnip yellow mosaic virus, and the polymerase of potato virus X are required for virus and/or RNA replication (7, 13, 14, 17, 21, 27, 44). In the case of brome mosaic virus (BMV), mutations designed to affect the helicase function of replicase protein 1a (homologous to AMV P1) inhibited viral negative-strand RNA synthesis in Saccharomyces cerevisiae (1).
FIG. 1.
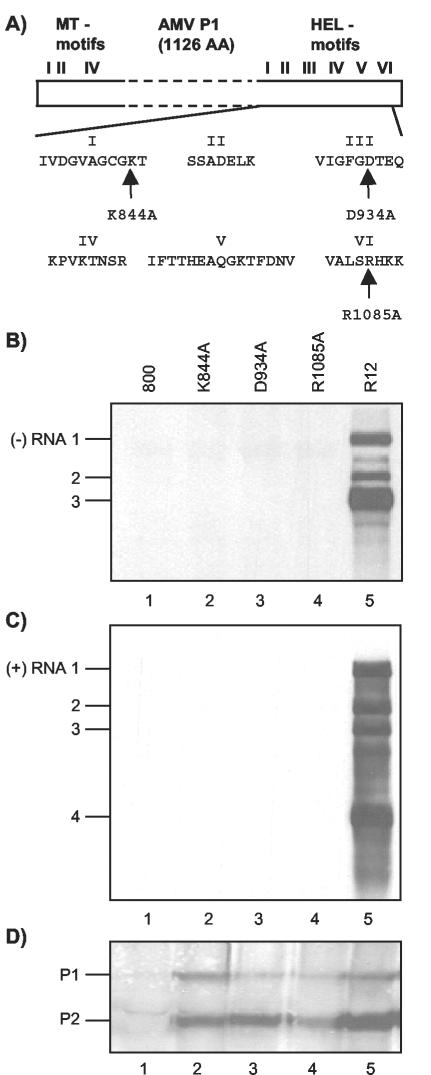
Mutations in RNA 1 affecting P1 helicase motifs interfere with viral RNA synthesis. (A) Schematic representation of P1 with N-terminal methyltransferase domain (MT) motifs and C-terminal helicase domain (HEL) motifs. The amino acids from helicase domain motifs I, III, and VI that are changed into alanine in mutants K844A, D934A, and R1085A, respectively, are indicated. (B to D) Analysis of the accumulation of viral negative-strand (−) RNAs (B), positive-strand (+) RNAs (C), and replicase proteins (D) in leaves infiltrated with a mixture of bacteria containing R12 and R3 constructs. The R3 construct expressed wt RNA 3 (lanes 1 to 5). The R12 construct expressed wt RNA 2 (lanes 2 to 5) and wt RNA 1 (lane 5) or RNA 1 encoding P1 with mutations in the helicase domain, as indicated above the lanes (lanes 2, 3, and 4). In leaves from which extracts were analyzed in lanes 1, the bacteria containing the R12 construct were replaced by bacteria containing the empty vector pMOG800 (800).Protein and RNA were extracted from the leaves 2 and 5 days p.i., respectively. RNA was analyzed by Northern blot hybridization using strand-specific probes (B and C). Protein was analyzed by Western blotting using antisera raised against P1 and P2 (D). The positions of RNAs 1, 2, 3, and 4 and proteins P1 and P2 are indicated on the left.
SF 1 helicases share seven conserved sequence motifs called I, Ia, and II to VI (4, 12, 18). Figure 1A depicts the AMV helicase motifs with the exception of motif Ia. Helicases of SF 1 and 2 share motifs I and II, which align with the ATP-binding and ATP-hydrolyzing Walker A and B motifs, respectively (4, 12). Mutation of the lysine residue of motif I (K844 in the case of AMV P1) is generally considered to disrupt ATP binding, thereby inhibiting ATP hydrolysis and duplex unwinding (4, 18). Motif III of the SF 1 DNA helicase PcrA of Bacillus stearothermophilus is thought to couple ATP hydrolysis to duplex unwinding (8). Several individual residues in this motif are essential for PcrA helicase activity (8, 9). In addition, two aspartate residues in PcrA motif III modulate intramolecular interactions (4). SF 1 helicase motif VI is thought to interact with ATP and function in the transition of energy from ATP hydrolysis to duplex unwinding (4). It has been shown that the Arg residue of PcrA motif VI (R1085 in AMV P1) interacts with ATP (40). Mutation of this residue to alanine strongly reduced both ATPase and duplex-unwinding activities of PcrA (33). Here, we have analyzed the role of the conserved residues K844, D934, and R1085 of AMV P1 in viral RNA replication.
Agrobacterium tumefaciens-mediated transient-expression assays have been used to express AMV RNAs and proteins from transferred-DNA (T-DNA) vectors in the leaves of Nicotiana benthamiana (42). Infiltration of the leaves results in agroinfection of a large proportion of the cells, and cell-to-cell movement is not required for viral negative- and positive-strand RNA accumulation. Thus, an advantage of the system is that wild-type (wt) or mutant viral RNAs and proteins can be expressed at relatively high levels without a requirement for replication of viral RNAs. Previously, agroinfiltration assays were used to show that mutations in the methyltransferase-like domain of P1 interfered with the synthesis of negative-strand AMV RNA (41). In the present study, we engineered mutations in RNA 1 affecting motifs I, III, and VI of the helicase-like domain of P1, mutants K844A, D934A, and R1085A, respectively (Fig. 1A). Mutant RNA 1 was expressed by agroinfiltration, together with RNAs 2 and 3 and/or wt replicase proteins, and the effects of the mutations on viral RNA replication were analyzed. The results prompted us to use the agroinfiltration assay to reinvestigate the phenotype of RNA 2 with a mutation in the sequence encoding the GDD motif of P2. Evidence was obtained that cis- and trans-acting functions of P1 and P2 are involved in the regulation of a coordinate replication of RNAs 1 and 2.
MATERIALS AND METHODS
DNA constructs.
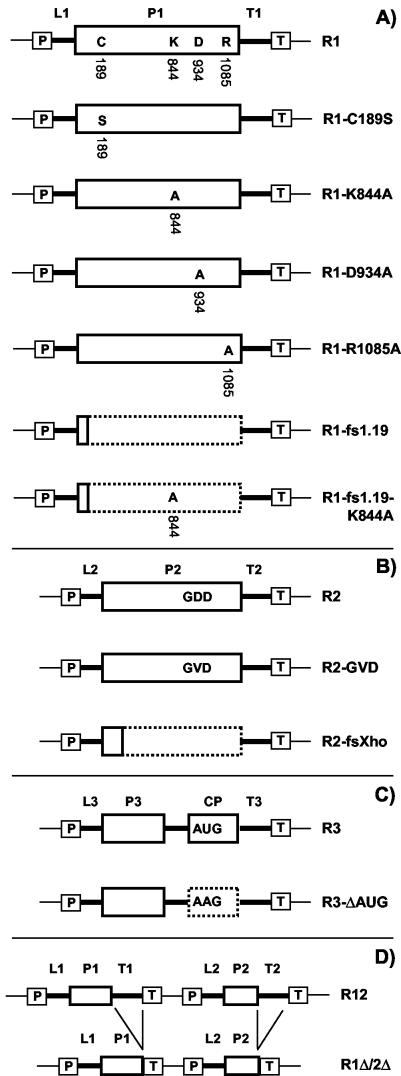
Figure 2 shows a diagrammatic illustration of all constructs used in this study. The construction of plasmids pBSR1, pBSR1-C189S, pMOGR1, pMOGR2, pMOGR12, pMOGR12-C189S, pMOGR3, and pMOGR1Δ/2Δ was described previously (42). pBSR1 contains a DNA copy of AMV RNA 1 cloned between the cauliflower mosaic virus (CaMV) 35S promoter and the terminator sequence of the nopaline synthase gene (Tnos). pMOG800 is the T-DNA vector used to express full-length DNA copies of RNA 1 (R1 [see Results]), RNA 2 (R2), and RNA 3 (R3) upon agroinfiltration. The empty vector is used as a negative control (42). pMOGR12 contains DNA copies of both RNAs 1 and 2. In pBSR1-C189S and pMOGR12-C189S, the DNA copy of RNA 1 encodes P1 carrying the C189S mutation. pMOGR1Δ/2Δ contains DNA copies of RNAs 1 and 2 with the 3′ untranslated regions deleted.
FIG.2.
Diagrammatic illustration of T-DNA constructs used in this study. Constructs R1 (A), R2 (B), and R3 (C) express wt RNAs 1, 2, and 3, respectively, whereas construct R12 (D) expresses wt RNAs 1 and 2. Mutant derivatives are shown below the corresponding wt constructs. Mutations in the DNA copy of RNA 1 were introduced both in the R1 construct (A) and in the R12 construct (diagrams not shown). The cDNA constructs are flanked by the CaMV 35S promoter (boxed P), nos terminator (boxed T), and T-DNA vector sequences (thin lines). AMV noncoding sequences are represented by thick lines. The 5′ untranslated regions of RNAs 1, 2, and 3 are labeled L1, L2, and L3, respectively. The 3′ untranslated regions of RNAs 1, 2, and 3 are labeled T1, T2, and T3, respectively. Sequences encoding the AMV proteins P1, P2, P3, and CP are indicated by solid-outline boxes. The dotted-outline boxes represent coding sequences that are untranslatable because of a frameshift or mutation of the initiation codon. See the text for further details.
Mutations were introduced using PCR-mediated site-directed mutagenesis. To introduce K844A, nucleotides 2365 to 2644 of a DNA copy of AMV RNA 1 were amplified using primers pCL14 (5′TACTTGGGTCGGACC3′) and pCo29 (5′GATATTGGTGGTAGCTCCGCAACCAG3′). Nucleotides 2618 to 3098 were amplified with primers pCo30 (5′CTGGTTGCGGAGCTACCACCAATATC3′) and pCo31 (5′CTTCGATAGATCTTAGTAC3′). The two fragments were fused by PCR with primers pCL14 and pCo31. The XbaI-ClaI fragment of the product of this PCR was exchanged with the corresponding fragment of pBSR1, yielding pBSR1-K844A. An AluI site was created by the introduction of the K844A mutation.
To introduce D934A, nucleotides 2365 to 2921 of the DNA copy of RNA 1 were amplified with primers pCL14 and pCo32 (5′GAATTTGTTCCGTGGCGCCAAAACCAATG3′). Nucleotides 2893 to 3098 were amplified with primers pCo33 (5′CATTGGTTTTGGCGCCACGGAACAAATTC3′) and pCo31. The two fragments were fused by PCR with primers pCL14 and pCo31. The ClaI-BglII fragment of the resulting PCR product was ligated to the BglII-ClaI fragment of the DNA copy of RNA 1 and cloned in pIC19H. Subsequently, the BglII-BglII fragment of this construct was exchanged with the corresponding fragment of pBSR1, yielding pBSR1-D934A. An HhaI site was created by the introduction of the D934A mutation.
To introduce R1085A, nucleotides 2365 to 3382 of the DNA copy of AMV RNA 1 were amplified by PCR with primers pCL14 and pCo34 (5′GGTAAAATATTTAAAAGTCTTCTTGTGCGCCGACAAGGCAAC3′). The BglII-SspI fragment of the product of this PCR was ligated to the SspI-SstI fragment of pBSR1 and cloned in pBluescript SK(+). The NcoI-SstI fragment of this construct was exchanged with the corresponding fragment of pBSR1, yielding pBSR1-R1085A. An HhaI site was created by the introduction of the R1085A mutation.
The fs1.19 frameshift mutation in RNA 1 was serendipitously obtained during PCR-mediated mutagenesis of a DNA copy of RNA 1. To disrupt an AluI site at position 203 in the DNA copy of RNA 1, a fragment of pBSR1 containing the 35S promoter and nucleotides 1 to 219 of the DNA copy of RNA 1 was amplified by PCR with primers pCo49 (5′GTAATACGACTCACTATAGGGC3′) and pCo50 (5′GTTGTGTCGTCTGCAGCCTGCTTTTCGACTACAC3′). The KpnI-PstI fragment of the product of this PCR was exchanged with the corresponding fragment of pBSR1. Upon sequencing of several clones, it was found that pBSR1-1.19 contained a deletion of nucleotide 195 of the DNA copy of RNA 1 and thus carried RNA 1 with a −1 frameshift mutation in the P1 reading frame. The construct was termed pBSR1-fs1.19. To obtain pBSR1-fs1.19-K844A, the XbaI-SstI fragment of pBSR1-K844A was exchanged with the corresponding fragment of pBSR1-fs1.19.
All PCR-derived fragments were sequenced (Baseclear, Leiden, The Netherlands). Subsequently, the R1 derivatives that were cloned in pBSR1 were transferred to pMOG800 or pMOGR2 using KpnI and SstI. Transfer to pMOG800 yielded the constructs pMOGR1-K844A, pMOGR1-D934A, pMOGR1-R1085A, pMOGR1-fs1.19, and pMOGR1-fs1.19-K844A. Transfer to pMOGR2 yielded the constructs pMOGR12-K844A, pMOGR12-D934A, pMOGR12-R1085A, and pMOGR12-fs1.19. In addition, cDNA1-C189S was transferred from pBSR1-C189S to pMOG800 using KpnI and SstI, yielding pMOGR1-C189S (42).
To obtain R2-GVD and R2-fsXho, pCA27T-GVD and pCA27T-fsXho were restricted with PvuII and SstI (39). The resulting fragments contained a DNA copy of RNA 2 with the relevant mutation, cloned between a 35S promoter and nos terminator. Each fragment was transferred to pMOG800, which had been restricted with SmaI and SstI. This yielded pMOGR2-GVD and pMOGR2-fsXho. To obtain pMOGR3-ΔAUG, the XhoI-SstI fragment of pAL3-CPΔAUG was exchanged with the corresponding fragment of pMOGR3 (37).
Agrobacterium-mediated transient expression.
All pMOG constructs were transformed to A. tumefaciens strain LBA4404 by electroporation. AMV RNAs were expressed from the T-DNA constructs by agroinfiltration of N. benthamiana leaves as described previously (42). Simultaneous expression of RNAs carried by different constructs was done by infiltration of mixtures of bacterial suspensions (41, 42). The suspensions were mixed in a 1:1(:1) ratio based on their optical densities at 600 nm (OD600). When two A. tumefaciens strains were coinfiltrated, the OD600 of the mixture was between 1.0 and 1.5, while the OD600 of each strain in the mixture was at least 0.5. When three A. tumefaciens strains were coinfiltrated, the OD600 of the mixture was between 1.2 and 1.5, and the OD600 of each strain in the mixture was at least 0.4. With the exception of the experiment with mutant R3-ΔAUG (see Fig. 4, lanes 6 to 10) and another experiment (see Fig. 5B), all experiments were repeated two to four times, and the results of a representative experiment are shown.
FIG. 4.
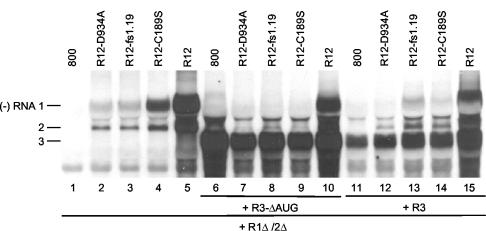
Competition among AMV RNAs for replication by a transiently expressed replicase. Leaves were agroinfiltrated with mixtures of bacteria containing constructs R1Δ/2Δ and R12 (lanes 2 to 5); constructs R1Δ/2Δ, R12, and R3-ΔAUG (lanes 7 to 10); or constructs R1Δ/2Δ, R12, and R3 (lanes 12 to 15). As negative controls, leaves were infiltrated with bacteria containing the empty vector pMOG800 (800) and R1Δ/2Δ (lane 1); pMOG800, R1Δ/2Δ, and R3-ΔAUG (lane 6); or pMOG800, R1Δ/2Δ, and R3 (lane 11). Construct R1Δ/2Δ expressed the wt proteins P1 and P2, R3 expressed wt RNA 3, R3-ΔAUG expressed RNA 3 with a mutation in the start codon of the CP gene, and the R12 constructs expressed wt RNA 2 and either wt RNA 1 (lanes 5, 10, and 15), RNA 1 with a mutation affecting a P1 helicase motif (lanes 2, 7, and 12), RNA 1 with a frameshift early in the P1 reading frame (lanes 3, 8, and 13), or RNA 1 with a mutation affecting the P1 methyltransferase domain (lanes 4, 9, and 14), as indicated above the lanes. Total RNA was extracted from the leaves 5 days p.i., and accumulation of negative-strand (−) RNAs was analyzed by Northern blot hybridization using strand-specific probes. The positions of RNAs 1, 2, and 3 are indicated on the left.
FIG. 5.
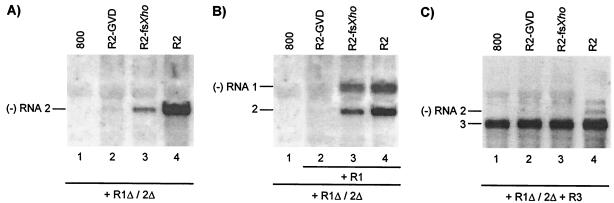
Analysis of cis- and trans-acting effects of mutations in RNA 2. Leaves were infiltrated with mixtures of bacteria, and total RNA was extracted from the leaves 2 (A and B) or 5 (C) days p.i. Accumulation of negative-strand (−) RNA was analyzed by Northern blot hybridization using strand-specific probes. The positions of RNAs 1, 2, and 3 are indicated on the left. (A) Coexpression of R1Δ/2Δ and R2 constructs (lanes 2, 3, and 4) or R1Δ/2Δ and the empty vector (lane 1). (B) Coexpression of R1Δ/2Δ, R2, and R1 constructs (lanes 2, 3, and 4) or R1Δ/2Δ and the empty vector (lane 1). (C) Coexpression of R1Δ/2Δ, R2, and R3 constructs (lanes 2, 3, and 4) or R1Δ/2Δ, R3, and the empty vector (lane 1). Construct R1Δ/2Δ expressed wt proteins P1 and P2. Constructs R1 and R3 expressed wt RNAs 1 and 3, respectively. Construct R2 expressed wt RNA 2 (lanes 4), RNA 2 with a mutation affecting the GDD motif in P2 (lanes 2), or RNA 2 with a frameshift early in the P2 reading frame (lanes 3).
Isolation and analysis of total RNA.
Total RNA was extracted from 250 mg of tissue 2 or 5 days after infiltration and analyzed by Northern blot hybridization as described previously (42). All Northern blot hybridizations were performed with nylon membranes (Roche). Five micrograms of RNA per slot was analyzed with digoxigenin-labeled riboprobes specific for minus-strand AMV RNAs 1, 2, and 3, whereas 0.05 μg of RNA was analyzed with digoxigenin-labeled riboprobes specific for plus-strand AMV RNAs 1, 2, and 3.
Protein analysis.
RdRp was isolated from ∼10 g of leaf tissue 2 days after infiltration. The leaves were homogenized, and large debris and nuclei were spun down. The supernatant was subsequently centrifuged at 30,000 × g for 20 min. The 30,000 × g pellet (P30) was resuspended in SOL buffer (80 mM Tris, pH 8.2, 15 mM MgCl2, 20% glycerol, 10 mM dithiothreitol, 50 nM aprotinin, 1 μM leupeptin, 1 μM pepstatin [Roche]). Samples from the P30 pellets corresponding to 50 mg of infiltrated tissue were loaded onto a Western blot. P1 and P2 were detected using antisera to the C-terminal amino acids 1100 to 1120 of P1 and the N terminus of P2 (36, 38). Western blotting was performed with Hybond-P polyvinylidene difluoride membranes (Amersham Pharmacia Biotech).
RESULTS
The AMV P1 helicase domain is required for negative-strand RNA synthesis.
Three mutations were individually introduced into helicase domain motifs I, III, and VI of the AMV replicase protein P1 (Fig. 1A). As outlined above, mutations K844A, D934A, and R1085A were expected to disrupt the putative helicase activity of P1 in different ways. To assess the role of this putative helicase in viral RNA replication, AMV RNA 1 encoding P1-K844A, P1-D934A, or P1-R1085A was expressed together with wt RNAs 2 and 3 in N. benthamiana leaves by agroinfiltration (41, 42). To that end, full-length mutant DNA copies of RNA 1 that had been cloned between a CaMV 35S promoter and nos terminator were transferred to the T region of the binary vector pMOGR2, which already carried a similar expression cassette containing a full-length DNA copy of RNA 2 (42). The resulting pMOGR12 derivatives were transformed to A. tumefaciens, and leaves were infiltrated with mixtures of bacteria containing pMOGR12-K844A, pMOGR12-D934A, or pMOGR12-R1085A and bacteria containing pMOGR3. The last vector carries a 35S expression cassette containing a full-length DNA copy of RNA 3 in its T region (42). Previously, it was shown that infiltration of N. benthamiana leaves with mixtures of bacteria containing the wt constructs pMOGR12 and pMOGR3 results in accumulation of both negative- and positive-strand AMV RNAs and CP at the level of a wt virus infection (41, 42). Hereafter, pMOGR1, pMOGR2, pMOGR3, and pMOGR12 are called R1, R2, R3, and R12 constructs, respectively. A diagrammatic illustration of the T-DNA constructs used in this study is shown in Fig. 2.
No negative- or positive-strand AMV RNAs were detectable in total-RNA extracts from leaves infiltrated with mixtures of bacteria containing R3 and one of the R12 derivatives encoding either P1-K844A, P1-D934A, or P1-R1085A (Fig. 1B and C, lanes 2 to 4). A mixture of bacteria containing R3 and the empty T-DNA vector pMOG800 was used as a negative control (Fig. 1B and C, lanes 1), while a mixture of bacteria containing the wt constructs R12 and R3 was used as a positive control (Fig. 1B and C, lanes 5). Previously, it was shown that the accumulation of positive- but not negative-strand AMV RNAs in agroinfiltrated leaves depends strongly on the accumulation of CP, which protects the RNAs from degradation (41, 42). While RNAs 1, 2, and 3 were transcribed from the T-DNA constructs by the host polymerase II, synthesis of the subgenomic CP-mRNA 4 depended solely on viral replicase complexes (41, 42). Therefore, the host pol.II transcripts of the A. tumefaciens T-DNAs were most likely degraded in the presence of P1 carrying one of the helicase domain mutations, since synthesis of RNA 4 was not supported (Fig. 1C, lanes 2 to 4).
Expression of RNAs 1 and 2 from the R12 constructs encoding P1 with a mutation in the helicase domain was verified by assessing the accumulation of their translation products. To that end, 30,000 × g membrane fractions from the infiltrated leaves were analyzed with antisera to P1 and P2. Both P1 and P2 accumulated to above background levels after the infiltration of leaves with mixtures of bacteria containing R3 and one of the R12 derivatives or wt R12 (Fig. 1D). Apparently, the mutations did not significantly affect the transient expression or stability of the replicase proteins. Moreover, Fig. 1D shows that transient expression of the AMV genome results in the accumulation of P1 and P2 at similar levels whether or not the viral RNAs are able to replicate (41, 42). Available evidence suggests that in the transient-expression system the synthesis of minus-strand RNA at a detectable level does not require de novo synthesis of plus-strand RNA by the viral RdRp (42). Thus, the results shown in Fig. 1B indicate that the three mutations in P1 affect functions of the protein in one or more steps leading to viral minus-strand RNA synthesis.
Complementation of RNA 1 mutants.
Previously, it was shown that transgenic tobacco plants expressing the P1 protein (P1 plants) supported the replication of AMV RNAs 2 and 3 in the absence of RNA 1 (35). Inoculation of P1 plants or protoplasts with the three genomic RNAs resulted in coreplication of RNA 1 with RNAs 2 and 3. However, frameshifts or deletions in the P1 reading frame abolished this coreplication, suggesting that replication of RNA 1 required the encoded P1 in cis (24, 39). In contrast, by using the agroinfiltration assay, we observed that replication of RNA 1 encoding point mutations in the methyltransferase domain of P1 (mutants P1-H100A and P1-C189S) could be complemented in trans by wt P1 (41). To analyze a possible complementation of the three helicase mutants by wt P1, leaves were infiltrated with a mixture of A. tumefaciens cultures harboring two different T-DNA vectors. One of these vectors (a derivative of construct R1) expressed the mutant RNA 1, and the other vector (construct R1Δ/2Δ) expressed the wt P1 and P2 proteins. RNAs 1 and 2 expressed from the R1Δ/2Δ construct are replication defective because they lack the 3′ untranslated regions (42) (Fig. 2D). Wt RNA 1 was efficiently transcribed into minus-strand RNA when coexpressed with the 3′-truncated RNAs 1 and 2 (Fig. 3A, lane 6). This transcription was mediated by P2 translated from the truncated RNA 2 and P1 translated either from the truncated RNA 1 or from wt RNA 1. As reported previously, RNA 1 encoding a mutation in the methyltransferase domain of P1 (mutant R1-C189S) (Fig. 2A) was also transcribed into minus-strand RNA in this system, although less efficiently than wt RNA 1 (41) (Fig. 3A, lane 5). As a replicase consisting of the mutant P1 and wt P2 is nonfunctional in minus-strand synthesis (41), transcription of the mutant RNA into minus strands was mediated by P1 and P2 expressed from the R1Δ/2Δ construct. Apparently, replication of RNA 1 with a mutation affecting the P1 methyltransferase domain can be partially complemented in trans by wt P1. However, no synthesis of minus-strand RNA was observed when RNA 1 encoding one of the helicase mutants of P1 was expressed together with the R1Δ/2Δ construct (Fig. 3A, lanes 2, 3, and 4). This indicates that replication of RNA 1 with a mutation affecting the P1 helicase domain cannot be complemented in trans.
FIG. 3.
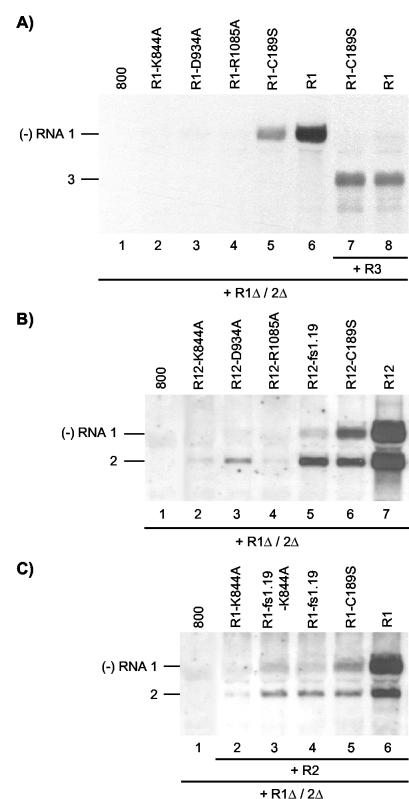
cis- and trans-acting effects of mutations in RNA 1 affecting P1 helicase motifs. Shown are Northern blot analyses of the accumulation of negative-strand (−) RNAs in agroinfiltrated leaves. (A) Coexpression of R1 and R1Δ/2Δ constructs (lanes 2 to 6) or R1, R1Δ/2Δ, and R3 constructs (lanes 7 and 8). The R1Δ/2Δ construct expressed the wt proteins P1 and P2, and the R3 construct expressed wt RNA 3. The R1 constructs expressed wt RNA 1 (lanes 6 and 8) or RNA 1 with mutations affecting helicase motifs (lanes 2, 3, and 4) or the methyltransferase domain (lanes 5 and 7) of P1, as indicated above the lanes. Total RNA was extracted from the leaves 2 (lanes 1 to 6) or 5 (lanes 7 and 8) days p.i. (B) Coexpression of R12 and R1Δ/2Δ constructs. The R12 construct expressed wt RNA 2 (lanes 2 to 7) and wt RNA 1 (lane 7), RNA 1 with mutations affecting helicase motifs (lanes 2, 3, and 4), or the methyltransferase domain (lane 6) of P1, or RNA 1 containing a frameshift early in the P1 reading frame (lane 5), as indicated above the lanes. Total RNA was extracted from the leaves 2 days p.i. (C) Coexpression of R1, R2, and R1Δ/2Δ constructs. The R2 construct expressed wt RNA 2 (lanes 2 to 6). The R1 construct expressed wt RNA 1 (lane 6), RNA 1 encoding P1 with a mutation in helicase motif I (lane 2), RNA 1 encoding the mutation in P1 helicase motif I as well as aframeshift early in the P1 reading frame (lane 3), RNA 1 with a frameshift early in the P1 reading frame (lane 4), or RNA 1 encoding P1 with a mutation in the methyltransferase domain (lane 5), as indicated above the lanes. (A to C) Total RNA was extracted from the leaves 2 days p.i. In leaves from which RNA extracts were analyzed in lanes 1, the bacteria containing the R1 (A and C) or R12 (B) constructs were replaced by bacteria containing the empty vector pMOG800 (800). The positions of RNAs 1, 2, and 3 are indicated on the left.
The P1 proteins with mutations in the putative methyltransferase or helicase domains could interfere partially or fully with the assembly of an active replicase from proteins expressed from the R1Δ/2Δ construct. To analyze this possibility, leaves were infiltrated with a mixture of A. tumefaciens cultures containing three T-DNA vectors: one with the R1Δ/2Δ construct expressing P1 and P2, one with an R3 construct expressing full-length RNA 3, and one with an R1 construct expressing wt RNA 1 or RNA 1 encoding P1 with a mutation in the methyltransferase domain (R1-C189S). Similar levels of minus-strand RNA 3 synthesis were observed in leaves infiltrated with mixtures of bacterial suspensions containing the mutant or wt RNA 1 construct (Fig. 3A, lanes 7 and 8). This level of minus-strand synthesis was not affected by omission of the bacteria containing the R1 constructs from the mixture used for agroinfiltration (result not shown). Apparently, expression of P1 with a mutation in the methyltransferase domain did not interfere with the assembly of an active replicase from wt proteins expressed from the R1Δ/2Δ construct. Similar experiments with the helicase mutants will be described below.
Mutant R1-C189S and wt RNA 1 were transcribed into minus-strand RNA in the absence of RNA 3 (Fig. 3A, lanes 5 and 6) but not in the presence of RNA 3 (Fig. 3A, lanes 7 and 8). RNA 3 may knock out the replication of RNA 1 in this system in a competition of the two templates for the replicase machinery induced by the R1Δ/2Δ construct.
Mutations in RNA 1 affect replication of RNA 2 in trans.
To analyze a possible trans-acting effect of mutations in RNA 1 on the replication of RNA 2, leaves were infiltrated with a mixture of bacteria containing the R1Δ/2Δ construct to express the wt replicase proteins and R12 constructs to express mutant RNA 1 and wt RNA 2. RNAs 1 and 2 expressed from a wt R12 construct induced the synthesis of similar levels of minus-strand RNAs 1 and 2 in the presence of the R1Δ/2Δ construct (Fig. 3B, lane 7) or in its absence (data not shown). The R12 construct expressing RNA 1 encoding the methyltransferase mutant P1-C189S did not induce minus-strand RNA synthesis in the absence of the R1Δ/2Δ construct but did so in the presence of the construct (41) (Fig. 3B, lane 6). As before (Fig. 3A, lane 5), synthesis of minus-strand RNA of the RNA 1 mutant was reduced, but a similar reduction was seen for the synthesis of minus-strand RNA 2 (Fig. 3B, lane 6). Expression of RNA 1 encoding the three P1 helicase mutants from R12 constructs in the presence of R1Δ/2Δ did not result in the synthesis of minus-strand RNA 1 (Fig. 3B, lanes 2, 3, and 4), similar to their expression from R1 constructs (Fig. 3A, lanes 2, 3 and 4). However, expression of the RNA 1 mutants also strongly inhibited transcription of wt RNA 2 into minus-strand RNA by the replicase expressed from the R1Δ/2Δ construct (Fig. 3B, lanes 2, 3, and 4). Expression of RNA 1 with a frameshift in codon 33 of the P1 reading frame (mutant R1-fs1.19) also resulted in reduced synthesis of both minus-strand RNAs 1 and 2 (Fig. 3B, lane 5) compared to the control (Fig. 3B, lane 7). Thus, the P1 proteins transiently expressed from RNA 1 with mutations affecting the methyltransferase or helicase domains of the protein may not be the sole determinants in the inhibition of RNA 2 replication in trans.
In the R12 constructs, the DNA copies of RNAs 1 and 2 are physically linked. To rule out the possibility that this linkage affected replication of RNA 2 by mutations in RNA 1, the two RNAs were expressed from separate R1 and R2 constructs. The synthesis of minus-strand RNAs 1 and 2 in leaves infiltrated with bacteria containing R1Δ/2Δ and wt R1 and R2 constructs is shown in Fig. 3C (lane 6). Similar to the results shown in Fig. 3B, introduction into the R1 construct of a mutation affecting the P1 methyltransferase (C189S) or helicase (K844A) domain or of a frameshift early in the P1 reading frame (fs1.19) resulted in a reduction in the synthesis of both minus-strand RNAs 1 and 2 (Fig. 3C, lanes 2, 4, and 5). Introduction of the fs1.19 frameshift into RNA 1 encoding the helicase mutation K844A slightly increased minus-strand synthesis of the RNA to the level of that of the mutant R1-fs1.19 (Fig. 3C, lane 3).
Competition among RNAs 1, 2, and 3 for replication machinery.
In addition to its structural role, CP has several functions in the AMV replication cycle. It is required to initiate infection by stimulating the translation of viral RNAs (25), it enhances the accumulation of plus-strand viral RNAs by protecting them from degradation (42), and it plays a role in cell-to-cell movement of the virus (29). To address a possible role of CP in the competition between AMV RNAs for a transiently expressed replicase, leaves were infiltrated with bacteria containing the R1Δ/2Δ construct for expression of the replicase, R12 constructs for expression of RNA 1 mutants, and R3 constructs for expression of wt RNA 3 or RNA 3 with a mutation in the start codon of the CP gene (mutant R3-ΔAUG) (Fig. 2C). RNA 3 with the ΔAUG mutation is defective in the expression of CP (37). The experiments were done with RNA 1 mutants encoding a mutation in the methyltransferase domain (C189S) or helicase domain (D934A) of P1 or with RNA 1 containing a frameshift early in the P1 gene (fs1.19) (Fig. 4). As a control, the synthesis of minus-strand RNAs induced by the wt and mutant R12 constructs in the absence of the R3 constructs was analyzed. As before (Fig. 3B), the three mutations reduced the accumulation of minus-strand RNAs 1 and 2 (Fig. 4, lanes 2, 3, and 4) compared to the wt control (Fig. 4, lane 5). In the experiments shown in Fig. 3B and 4, the RNAs were isolated 2 and 5 days postinfiltration (p.i.) of the leaves, respectively (see the figure legends). Apparently, at 2 days p.i., minus-strand accumulation of mutant R1-D934A is lower than minus-strand accumulation of mutant R1-fs1.19 (Fig. 3B, lanes 3 and 5), whereas at 5 days p.i., similar amounts of minus-strand RNAs of these mutants were accumulated (Fig. 4, lanes 2 and 3).
Coexpression of the R12 and R1Δ/2Δ constructs, together with the R3 constructs, resulted in synthesis of minus-strand RNA corresponding to mutant RNA 3 (Fig. 4, lanes 6 to 10) or wt RNA 3 (Fig. 4, lanes 11 to 15). The synthesis of mutant and wt minus-strand RNA 3 was accompanied by a significant reduction in the synthesis of minus-strand RNAs 1 and 2 (Fig. 4, lanes 6 to 15) compared to the controls (Fig. 4, lanes 2 to 5). This indicates that CP plays no role in the preferential replication of RNA 3 by the replicase expressed from the R1Δ/2Δ construct. Apparently, RNA 3 outcompetes not only replication of RNA 1 (Fig. 3A, lanes 7 and 8) but also replication of RNA 2 (Fig. 4). Moreover, the data showed that P1 with a mutation in the helicase domain (Fig. 4, lanes 7 and 12) or the N-terminal peptide potentially expressed from RNA 1 with the frameshift mutation (Fig. 4, lanes 8 and 13) did not interfere with assembly of the replicase that replicates RNA 3. The earlier conclusion that the methyltransferase mutant (P1-C189S) did not interfere with assembly of the replicase (Fig. 3A, lane 7) was confirmed by the results shown in Fig. 4 (lanes 9 and 14).
Functions involved in replication of RNA 2.
Transgenic tobacco plants expressing the P2 protein (P2 plants) support the replication of AMV RNAs 1 and 3 in the absence of RNA 2 (35). The transgenically expressed P2 did not interfere with the coreplication of wt RNA 2 with RNAs 1 and 3 but did not support coreplication of RNA 2 with a mutation changing the GDD sequence in the polymerase domain of the P2 protein into a GVD sequence (mutant GVD) (Fig. 2B) (39). These data indicated that replication of RNA 2 required the encoded P2 in cis. Here, we investigated a possible complementation of the replication of the RNA 2 mutants in trans by the transiently expressed replicase expressed from the R1Δ/2Δ construct. Wt RNA 2 expressed from the R2 construct was efficiently transcribed into minus-strand RNA when coexpressed with the R1Δ/2Δ construct (Fig. 5A, lane 4). A frameshift in codon 70 of the P2 gene (mutant fsXho) strongly reduced minus-strand synthesis of the RNA (Fig. 5A, lane 3). In addition, minus-strand synthesis of RNA 2 encoding P2 with the GVD mutation was virtually abolished (Fig. 5A, lane 2). Previously, it was shown that both mutations abolish infectivity of the virus (39). Apparently, complementation of the replication of these mutants in trans by wt P2 occurs only at a low level or not at all.
To see if the mutations in RNA 2 affected the replication of RNA 1 in trans, leaves were infiltrated with mixtures of bacteria containing the R1Δ/2Δ construct, the wt R1 construct, and R2 constructs expressing wt or mutant RNA 2. Wt RNAs expressed from the R1 and R2 constructs induced the synthesis of minus-strand RNAs 1 and 2 (Fig. 5B, lane 4). Introduction of the fsXho frameshift mutation into RNA 2 resulted in a small but significant reduction in the synthesis of both minus-strand RNAs 1 and 2 (Fig. 5B, lane 3). However, introduction of the GVD mutant coding sequence into RNA 2 virtually abolished the synthesis of minus-strand RNAs 1 and 2 (Fig. 5B, lane 2). Thus, mutations in RNA 2 interfered in trans with the replication of RNA 1.
Infiltration of leaves with bacteria containing the R1Δ/2Δ, R3, and wt or mutant R2 constructs showed that proteins potentially expressed from R2-GVD and R2-fsXho did not interfere with the assembly of a replicase that replicated RNA 3 expressed from the R3 construct (Fig. 5C, lanes 2 and 3). Although wt RNA 2 and RNA 2 with the fsXho mutation induced minus-strand RNA 2 synthesis in the absence of RNA 3 (Fig. 5A, lanes 3 and 4), no minus-strand synthesis of these RNAs was observed when RNA 3 was coexpressed (Fig. 5C, lanes 3 and 4). This confirms that neither RNA 1 nor RNA 2 is able to compete with RNA 3 for replication by the transiently expressed replicase.
DISCUSSION
Role of the helicase-like domain in AMV RNA replication.
Replication-defective RNAs 1 and 2 expressed from a T-DNA vector accumulate at relatively high levels when CP is coexpressed from RNA 3. These plus-strand transcripts may accumulate to levels of up to 50% of the levels of RNAs 1 and 2 produced during a wt AMV infection (42) and should provide ample templates for minus-strand RNA synthesis by functional replicases. Transcripts encoding mutations in motif I, III, or VI of the helicase-like domain of P1 were efficiently translated into replicase proteins (Fig. 1D), but the mutant replicases did not transcribe the RNA 1, 2, or 3 transcripts into minus-strand RNAs (Fig. 1B). As at least RNAs 2 and 3 were functional templates in this experiment, we conclude that the mutant replicases are defective in minus-strand RNA synthesis and that, as a consequence, there is no plus-strand RNA synthesis (Fig. 1C). Mutations corresponding to K844A in the helicase or helicase-like domains of a number of other viruses have been shown to interfere with replication of the virus or of the viral RNA (1, 7, 14, 16, 21, 23, 27, 44). For several viruses, a mutational analysis revealed a correlation between virus viability and NTPase or duplex-unwinding activities of the viral helicase protein (10, 15, 16, 23, 27). Similar to AMV P1, the helicase domains of BVDV NS3 and BMV 1a were found to be required for negative-strand RNA synthesis (1, 16). Mutations in the BMV helicase domain appeared to affect the function of 1a in the recruitment of viral RNA templates to replication complexes (1). Moreover, the helicases of BMV, HCV, and TMV are believed to act as oligomers, and the helicase domains may be involved in oligomerization of replicase proteins (1, 13, 14, 20). By analogy, the mutations in the helicase domain of AMV P1 may affect functions of the P1 protein preceding initiation of minus-strand RNA synthesis.
cis-acting coding functions in AMV RNAs 1 and 2.
RNAs 1, 2, and 3 are able to replicate separately in the presence of a wt replicase that is transiently expressed from the R1Δ/2Δ construct (Fig. 3A, lane 6; Fig. 5A, lane 4, and C, lane 1). Mutations in the P3 or CP gene did not affect minus-strand RNA 3 synthesis by AMV replicase expressed in transgenic plants (24) or in agroinfiltrated leaves (Fig. 4). However, mutations in the coding sequences of RNAs 1 and 2 severely reduced or abolished replication of these RNAs in trans by the transiently expressed wt replicase (Fig. 3A, lanes 2 to 5, and 5A, lanes 2 and 3). As the mutations did not affect coreplication of RNA 3, the mutant proteins apparently did not interfere with the assembly of the transiently expressed replicase. One could speculate that replication of RNA 1 requires the helicase function in cis (Fig. 3A, lanes 2, 3, and 4) whereas mutation of the methyltransferase function of P1 can be partially complemented in trans (Fig. 3A, lane 5). However, the frameshift mutant fs1.19 does not express the methyltransferase or helicase domain of P1, and replication of the mutant RNA is complemented in trans at a low but detectable level (Fig. 3B, lane 5). Mutagenesis of the start codons of the P1 and P2 genes will be required to analyze possible cis-acting negative effects of mutant P1 and mutant P2, or N-terminal peptides of these proteins, on the replication of RNAs 1 and 2, respectively.
Silent mutations in the region of RNA 2 encoding the P2 GDD sequence did not affect RNA 2 replication (39). Thus, we propose that the GVD mutation interferes with RNA 2 replication by affecting the function of the P2 protein rather than by affecting the promoter for minus-strand RNA synthesis in RNA 2. The poor complementation by wt replicase proteins of minus-strand synthesis of RNAs 1 and 2 with mutations in the coding regions of P1 and P2 indicates that one or more functions of these proteins are required in cis for replication. The TMV 126-kDa protein (equivalent to AMV P1) has been shown to be required in cis for TMV RNA replication (21). Moreover, replication of turnip yellow mosaic virus RNA with mutations in the helicase or polymerase coding regions was poorly complemented in trans (44). In contrast, replication of BMV RNA 2 mutants with a frameshift or deletion in the 2a gene could be efficiently complemented by wt 2a in trans (5).
Coordinate replication of AMV RNAs 1 and 2.
RNA 1 is able to recruit P2 expressed from the R1Δ/2Δ construct for its replication (Fig. 3A, lane 6), and similarly, RNA 2 recruits P1 expressed from this construct (Fig. 5A, lane 4). However, when RNAs 1 and 2 were replicating together in cells in which R1Δ/2Δ was expressed, mutations in RNA 1 reduced or completely abolished the ability of RNA 2 to replicate by recruitment of wt P1 expressed from R1Δ/2Δ (Fig. 3B and C). However, the mutations in RNA 1 did not interfere with the replication of RNA 3 by the transiently expressed replicase (Fig. 4). Also, the GVD mutation encoded by RNA 2 fully blocked the replication of RNA 1 in the presence of wt P2 expressed from R1Δ/2Δ (Fig. 5B, lane 2). This points to a mechanism that coordinates the replication of RNAs 1 and 2 when both genome segments are present in one cell. BMV 1a is present in 25-fold excess over BMV 2a in membrane structures with replicase activity (30). The TMV replicase contains the 126-kDa protein (equivalent to AMV P1 and BMV 1a) and the 183-kDa readthrough protein with the polymerase domain (equivalent to AMV P2 and BMV 2a). The 126-kDa protein is present in infected cells in 10-fold excess over the 183-kDa protein, but it was shown that both proteins coimmunoprecipitate in a 1:1 ratio (43). In addition to their roles in RNA replication, BMV 1a and the TMV 126-kDa protein may perform other functions requiring tight regulation of their synthesis.
Recently, a cis-acting mechanism was uncovered for the recruitment of BMV RNA 2 to replication complexes (6) that is alternative to the box B-dependent pathway acting in trans (5, 30). Recruitment of RNA 2 by this mechanism requires translation of the region of the encoded 2a protein that interacts with the helicase domain of 1a (6). Similarly, the requirement for AMV P1 and P2 in cis and interactions between the helicase domain of P1 and the N-terminal region of P2 (36) may play a role in the coordination of the replication of RNAs 1 and 2. However, the results with the frameshift mutants indicate that other factors may play a role as well. RNA 3 is exclusively replicated in trans by P1 and P2, and mutations in RNA 1 or 2 did not affect RNA 3 replication in the experiments shown in Fig. 3, 4, and 5.
Competition between AMV RNAs for the viral replicase.
Coexpression of RNAs 1 and 3 or RNAs 2 and 3 with the R1Δ/2Δ construct resulted in the selective replication of RNA 3 only (Fig. 3A, lane 8, and 5C, lane 4). Similarly, inoculation of transgenic tobacco plants expressing the P1 and P2 proteins (P12 plants) with RNAs 1, 2, and 3 resulted in a rapid loss of RNAs 1 and 2 from the progeny virus (35). This points to a mechanism that permits the virus to optimize its replication strategy by replicating only those RNAs that are essential for producing progeny that is infectious to the type of cell in which it was produced. Previously, we reported that wt RNA 1 coreplicated with RNAs 2 and 3 in P1 plants or protoplasts whereas several RNA 1 mutants did not coreplicate in P1 plants, although these mutants were fully viable in nontransgenic plants (24, 39). Similar results were observed with the replication of mutant RNA 2 in P2 plants or protoplasts (24, 39). This indicates that the replicase not only selects the genomic RNAs that are essential for the production of progeny but even selects the fittest template from a population of variants of a given genome segment. Such a selectivity of the replicase may serve as a bottleneck in the passage of mutations to the progeny virus.
In summary, our results show that conserved motifs in the helicase-like domain of P1 act in cis in one or more steps leading to AMV minus-strand RNA synthesis and point to the existence of mechanisms that coordinate the synthesis of RNAs 1 and 2 and provide selectivity to the replicase in the use of viral template RNAs.
REFERENCES
- 1.Ahola, T., J. A. Den Boon, and P. Ahlquist. 2000. Helicase and capping enzyme active site mutations in brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping. J. Virol. 74:8803-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bautista, E. M., K. S. Faaberg, D. Mickelson, and E. D. McGruder. 2002. Functional properties of the predicted helicase of porcine reproductive and respiratory syndrome virus. Virology 298:258-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bol, J. F. 2003. Alfalfa mosaic virus: coat protein-dependent initiation of infection. Mol. Plant Pathol. 4:1-8. [DOI] [PubMed] [Google Scholar]
- 4.Caruthers, J. M., and D. B. McKay. 2002. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12:123-133. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J., A. Noueiry, and P. Ahlquist. 2001. Brome mosaic virus protein 1a recruits viral RNA 2 to RNA replication through a 5′ proximal RNA 2 signal. J. Virol. 75:3207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, J., A. Noueiry, and P. Ahlquist. 2003. An alternative pathway for recruiting template RNA to the brome mosaic virus RNA replication complex. J. Virol. 77:2568-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davenport, G. F., and D. C. Baulcombe. 1997. Mutation of the GKS motif of the RNA-dependent RNA polymerase from potato virus X disables or eliminates virus replication. J. Gen. Virol. 78:1247-1251. [DOI] [PubMed] [Google Scholar]
- 8.Dillingham, M. S., P. Soultanas, and D. B. Wigley. 1999. Site-directed mutagenesis of motif III in PcrA helicase reveals a role in coupling ATP hydrolysis to strand separation. Nucleic Acids Res. 27:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dillingham, M. S., P. Soultanas, P. Wiley, M. R. Webb, and D. B. Wigley. 2001. Defining the roles of individual residues in the single-stranded DNA binding site of PcrA helicase. Proc. Natl. Acad. Sci. USA 98:8381-8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernández, A., H. S. Guo, P. Sáenz, L. Simón-Buela, M. Gómez de Cedrón, and J. A. García. 1997. The motif V of plum pox potyvirus Cl RNA helicase is involved in NTP hydrolysis and is essential for virus RNA replication. Nucleic Acids Res. 25:4474-4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez de Cedrón, M., N. Ehsani, M. L. Mikkola, J. A. García, and L. Kääriäinen. 1999. RNA helicase activity of Semliki Forest virus replicase protein nsP2. FEBS Lett. 448:19-22. [DOI] [PubMed] [Google Scholar]
- 12.Gorbalenya, A. E., and E. V. Koonin. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3:419-429. [Google Scholar]
- 13.Goregaoker, S. P., and J. N. Culver. 2003. Oligomerization and activity of the helicase domain of the tobacco mosaic virus 126- and 183-kilodalton replicase proteins. J. Virol. 77:3549-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goregaoker, S. P., D. J. Lewandowski, and J. N. Culver. 2001. Identification and functional analysis of an interaction between domains of the 126/183-kDa replicase-associated proteins of tobacco mosaic virus. Virology 282:320-328. [DOI] [PubMed] [Google Scholar]
- 15.Grassmann, C. W., O. Isken, and S.-E. Behrens. 1999. Assignment of the multifunctional NS3 protein of bovine viral diarrhea virus during RNA replication: an in vivo and in vitro study. J. Virol. 73:9196-9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu, B., C. Liu, J. Lin-Goerke, D. R. Maley, L. L. Gutshall, C. A. Feltenberger, and A. M. Del Vecchio. 2000. The RNA helicase and nucleotide triphosphatase activities of the bovine viral diarrhea virus NS3 protein are essential for viral replication. J. Virol. 74:1794-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadaré, G., C. David, and A.-L. Haenni. 1996. ATPase, GTPase, and RNA binding activities associated with the 206-kilodalton protein of turnip yellow mosaic virus. J. Virol. 70:8169-8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadaré, G., and A.-L. Haenni. 1997. Virus-encoded RNA helicases. J. Virol. 71:2583-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levin, M. K., and S. S. Patel. 1999. The helicase from hepatitis C virus is active as an oligomer. J. Biol. Chem. 274:31839-31846. [DOI] [PubMed] [Google Scholar]
- 21.Lewandowski, D. J., and W. O. Dawson. 2000. Functions of the 126- and 183-kDa proteins of tobacco mosaic virus. Virology 271:90-98. [DOI] [PubMed] [Google Scholar]
- 22.Locatelli, G. A., S. Spadari, and G. Maga. 2002. Hepatitis C virus NS3 ATPase/helicase: an ATP switch regulates the cooperativity among the different substrate binding sites. Biochemistry 41:10332-10342. [DOI] [PubMed] [Google Scholar]
- 23.Matusan, A. E., M. J. Pryor, A. D. Davidson, and P. J. Wright. 2001. Mutagenesis of the Dengue virus type 2 NS3 protein within and outside helicase motifs: effects on enzyme activity and virus replication. J. Virol. 75:9633-9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neeleman, L., and J. F. Bol. 1999. cis-acting functions of alfalfa mosaic virus proteins involved in replication and encapsidation of viral RNA. Virology 254:324-333. [DOI] [PubMed] [Google Scholar]
- 25.Neeleman, L., R. C. L. Olsthoorn, H. J. M. Linthorst, and J. F. Bol. 2001. Translation of a nonpolyadenylated viral RNA is enhanced by binding of viral coat protein or polyadenylation of the RNA. Proc. Natl. Acad. Sci. USA 98:14286-14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Reilly, E. K., and C. C. Kao. 1998. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 252:287-303. [DOI] [PubMed] [Google Scholar]
- 27.Rikkonen, M. 1996. Functional significance of the nuclear-targeting and NTP-binding motifs of Semliki Forest virus nonstructural protein nsP2. Virology 218:352-361. [DOI] [PubMed] [Google Scholar]
- 28.Rozanov, M. N., E. V. Koonin, and A. E. Gorbalenya. 1992. Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J. Gen. Virol. 73:2129-2134. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez-Navarro, J. A., and J. F. Bol. 2001. Role of the alfalfa mosaic virus movement protein and coat protein in virus transport. Mol. Plant-Microbe Interact. 14:1051-1062. [DOI] [PubMed] [Google Scholar]
- 30.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. Den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 31.Seybert, A., A. Hegyi, S. G. Siddell, and J. Ziebuhr. 2000. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA 6:1056-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seybert, A., L. C. Van Dinten, E. J. Snijder, and J. Ziebuhr. 2000. Biochemical characterization of the equine arteritis virus helicase suggests a close functional relationship between arterivirus and coronavirus helicases. J. Virol. 74:9586-9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soultanas, P., M. S. Dillingham, S. S. Velankar, and D. B. Wigley. 1999. DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase. J. Mol. Biol. 290:137-148. [DOI] [PubMed] [Google Scholar]
- 34.Tai, C.-L., W.-C. Pan, S.-H. Liaw, U.-C. Yang, L.-H. Hwang, and D.-S. Chen. 2001. Structure-based mutational analysis of the hepatitis C virus NS3 helicase. J. Virol. 75:8289-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taschner, P. E. M., A. C. van der Kuyl, L. Neeleman, and J. F. Bol. 1991. Replication of an incomplete alfalfa mosaic virus genome in plants transformed with viral replicase genes. Virology 181:445-450. [DOI] [PubMed] [Google Scholar]
- 36.Van Der Heijden, M. W., J. E. Carette, P. J. Reinhoud, A. Haegi, and J. F. Bol. 2001. Alfalfa mosaic virus replicase proteins P1 and P2 interact and colocalize at the vacuolar membrane. J. Virol. 75:1879-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van der Vossen, E. A. G., L. Neeleman, and J. F. Bol. 1994. Early and late functions of alfalfa mosaic virus coat protein can be mutated separately. Virology 202:891-903. [DOI] [PubMed] [Google Scholar]
- 38.Van Pelt-Heerschap, H. 1987. Immunochemical analysis of the alfalfa mosaic virus gene products. Ph.D. thesis. Leiden University, Leiden, The Netherlands.
- 39.Van Rossum, C. M. A., M. L. Garcia, and J. F. Bol. 1996. Accumulation of alfalfa mosaic virus RNAs 1 and 2 requires the encoded proteins in cis. J. Virol. 70:5100-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Velankar, S. S., P. Soultanas, M. S. Dillingham, H. S. Subramanya, and D. B. Wigley. 1999. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97:75-84. [DOI] [PubMed] [Google Scholar]
- 41.Vlot, A. C., A. Menard, and J. F. Bol. 2002. Role of the alfalfa mosaic virus methyltransferase-like domain in negative-strand RNA synthesis. J. Virol. 76:11321-11328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vlot, A. C., L. Neeleman, H. J. M. Linthorst, and J. F. Bol. 2001. Role of the 3′-untranslated regions of alfalfa mosaic virus RNAs in the formation of a transiently expressed replicase in plants and in the assembly of virions. J. Virol. 75:6440-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe, T., A. Honda, A. Iwata, S. Ueda, T. Hibi, and A. Ishihama. 1999. Isolation from tobacco mosaic virus-infected tobacco of a solubilized template-specific RNA-dependent RNA polymerase containing a 126K/183K protein heterodimer. J. Virol. 73:2633-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiland, J. J., and T. W. Dreher. 1993. cis-preferential replication of the turnip yellow mosaic virus RNA genome. Proc. Natl. Acad. Sci. USA 90:6095-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]