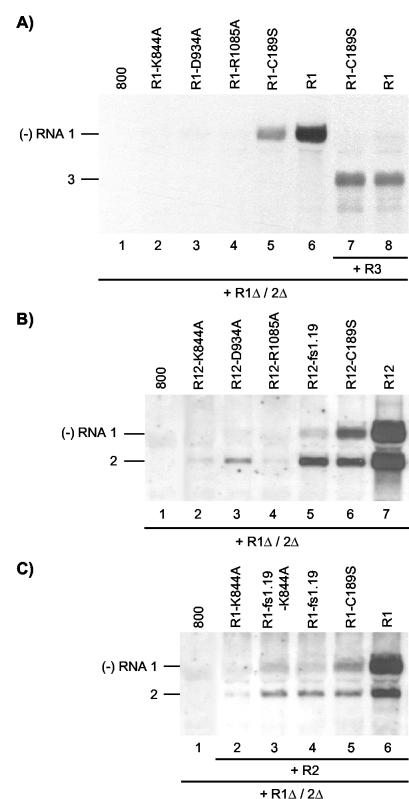
FIG. 3.
cis- and trans-acting effects of mutations in RNA 1 affecting P1 helicase motifs. Shown are Northern blot analyses of the accumulation of negative-strand (−) RNAs in agroinfiltrated leaves. (A) Coexpression of R1 and R1Δ/2Δ constructs (lanes 2 to 6) or R1, R1Δ/2Δ, and R3 constructs (lanes 7 and 8). The R1Δ/2Δ construct expressed the wt proteins P1 and P2, and the R3 construct expressed wt RNA 3. The R1 constructs expressed wt RNA 1 (lanes 6 and 8) or RNA 1 with mutations affecting helicase motifs (lanes 2, 3, and 4) or the methyltransferase domain (lanes 5 and 7) of P1, as indicated above the lanes. Total RNA was extracted from the leaves 2 (lanes 1 to 6) or 5 (lanes 7 and 8) days p.i. (B) Coexpression of R12 and R1Δ/2Δ constructs. The R12 construct expressed wt RNA 2 (lanes 2 to 7) and wt RNA 1 (lane 7), RNA 1 with mutations affecting helicase motifs (lanes 2, 3, and 4), or the methyltransferase domain (lane 6) of P1, or RNA 1 containing a frameshift early in the P1 reading frame (lane 5), as indicated above the lanes. Total RNA was extracted from the leaves 2 days p.i. (C) Coexpression of R1, R2, and R1Δ/2Δ constructs. The R2 construct expressed wt RNA 2 (lanes 2 to 6). The R1 construct expressed wt RNA 1 (lane 6), RNA 1 encoding P1 with a mutation in helicase motif I (lane 2), RNA 1 encoding the mutation in P1 helicase motif I as well as aframeshift early in the P1 reading frame (lane 3), RNA 1 with a frameshift early in the P1 reading frame (lane 4), or RNA 1 encoding P1 with a mutation in the methyltransferase domain (lane 5), as indicated above the lanes. (A to C) Total RNA was extracted from the leaves 2 days p.i. In leaves from which RNA extracts were analyzed in lanes 1, the bacteria containing the R1 (A and C) or R12 (B) constructs were replaced by bacteria containing the empty vector pMOG800 (800). The positions of RNAs 1, 2, and 3 are indicated on the left.