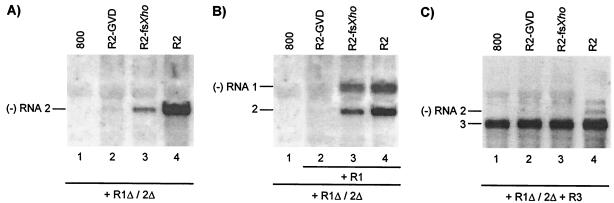
FIG. 5.
Analysis of cis- and trans-acting effects of mutations in RNA 2. Leaves were infiltrated with mixtures of bacteria, and total RNA was extracted from the leaves 2 (A and B) or 5 (C) days p.i. Accumulation of negative-strand (−) RNA was analyzed by Northern blot hybridization using strand-specific probes. The positions of RNAs 1, 2, and 3 are indicated on the left. (A) Coexpression of R1Δ/2Δ and R2 constructs (lanes 2, 3, and 4) or R1Δ/2Δ and the empty vector (lane 1). (B) Coexpression of R1Δ/2Δ, R2, and R1 constructs (lanes 2, 3, and 4) or R1Δ/2Δ and the empty vector (lane 1). (C) Coexpression of R1Δ/2Δ, R2, and R3 constructs (lanes 2, 3, and 4) or R1Δ/2Δ, R3, and the empty vector (lane 1). Construct R1Δ/2Δ expressed wt proteins P1 and P2. Constructs R1 and R3 expressed wt RNAs 1 and 3, respectively. Construct R2 expressed wt RNA 2 (lanes 4), RNA 2 with a mutation affecting the GDD motif in P2 (lanes 2), or RNA 2 with a frameshift early in the P2 reading frame (lanes 3).