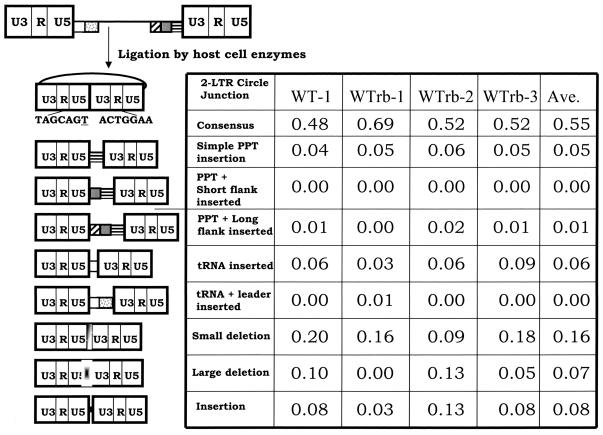
FIG. 2.
2-LTR circle junctions obtained after infecting cells with wild-type virus. The top of the figure shows the linear viral DNA that is the product of reverse transcription. The first column shows the results obtained with the original vector pNLNgoMIVR-E-.HSA. The wild-type PPT sequence was reconstructed by using the same BspMI mutagenesis protocol used to construct the PPT mutants, and the reconstructed vector was sequenced to show, it was identical to the original vector. Three independent experiments were performed that using a rebuilt vector containing the wild-type PPT; these are designated WTrb in the top of the columns. The pbs is indicated by a white box, the leader sequence downstream of the pbs is shown with gray dots, the PPT is a box with black horizontal bars, the U-tract is a gray box, and the sequences immediately upstream of the U-tract are indicated by the box with diagonal bars. The linear viral DNA can be ligated by cellular enzymes in the nucleus of an infected cell to form a 2-LTR circle. A consensus circle junction is shown at the top. Different categories of aberrant 2-LTR circle junctions are shown. The underlined “T” is derived from the riboA located at the end of the negative strand; it is derived from the tRNALys3 primer used by HIV-1 RT to initiate negative-strand DNA synthesis. The numbers of samples analyzed were 80 for the parental vector and 64, 64, and 77, respectively, for the three experiments with the rebuilt vector.