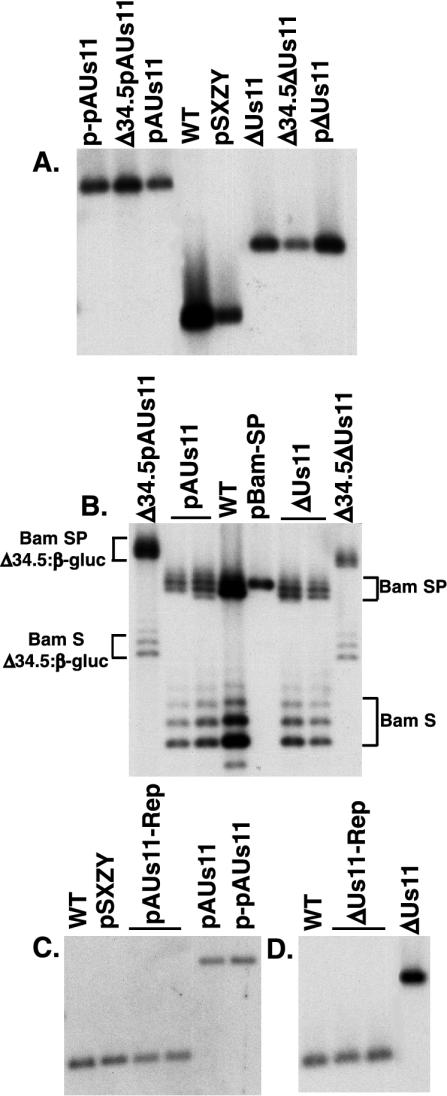
FIG. 2.
Physical structure of mutant loci in HSV-1 recombinants. (A) EcoNI-digested viral and plasmid DNAs were fractionated by electrophoresis in agarose gels, transferred to nylon membranes, and incubated with a 32P-labeled EcoNI-NcoI DNA probe that detects the Us10-to-Us12 region. The washed membrane was subsequently exposed to Kodak XAR film. (B) Viral and plasmid DNAs were digested with BamHI, fractionated by agarose gel electrophoresis, transferred to nylon membranes, and incubated with a 32P-labeled BamHI-BstXI probe derived from the BamHI SP fragment DNA probe. The washed filter was exposed to Kodak XAR film. This probe hybridizes to a segment adjacent to the γ134.5 ORF in the BamHI S and SP fragments, and therefore each fragment contains either one copy of the γ134.5 gene or, for the γ134.5 deletion mutants, one copy of the β-glucuronidase (β-gluc) gene. Heterogeneity in the mobility of the BamHI S and SP fragments is due to natural variations in the length of a repetitive sequence element. (C and D) EcoNI-digested viral and plasmid DNAs were separated in agarose gels, transferred to nylon membranes, and incubated with a 32P-labeled NcoI-PflMI DNA probe that detects the Us10-to-Us12 region. After being washed, the filter was exposed to Kodak XAR film.