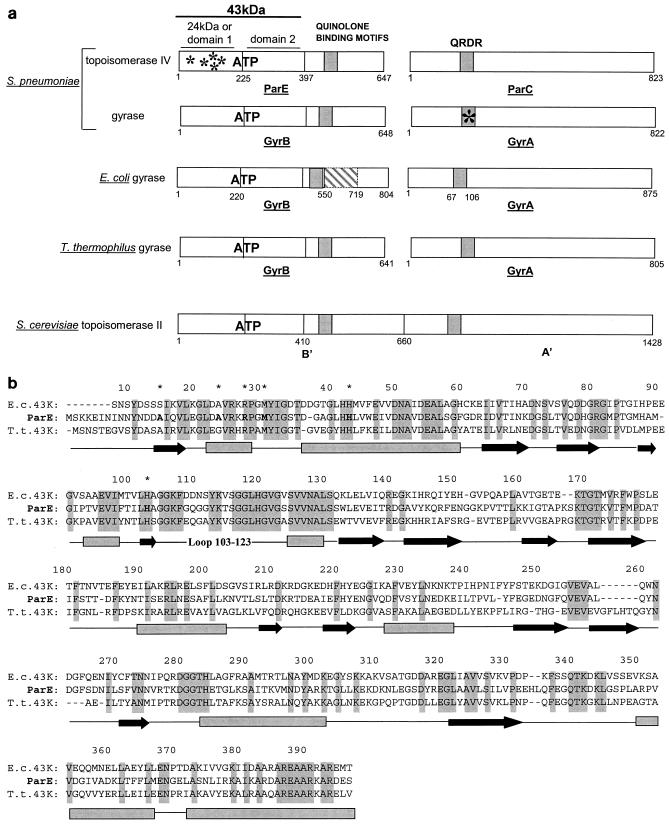
FIG. 2.
Comparison of different topoisomerases and of the 43-kDa domains of GyrB and ParE. (a) Schematic representation of gyrase and topoisomerase IV subunits. The bacterial topoisomerases are composed of two subunits associated in an A2B2 tetramer (DNA gyrase) and a C2E2 tetramer (topoisomerase IV). The eukaryotic topoisomerase II contains in the same polypeptide sequences homologous to the ParE/GyrB (B′) and ParC/GyrA (A′) bacterial subunits. All topoisomerases contain an ATP binding site lying in the first 43-kDa domain of the ParE/GyrB subunit.E. coli gyrase B contains a large insertion (cross-hatched box) between positions 550 and 719. Quinolone binding motifs lie mainly in the N-terminal region of ParC/GyrA (QRDR) as well as in the C-terminal part of ParE/GyrB. In this study, S. pneumoniae mutations in the 24-kDa N terminus of ParE were found in quinolone-resistant strains associated with the Ser81Phe mutation in the QRDR of GyrA (asterisks). (b) Alignment of the 43-kDa domain sequences of E. coli (E. c.) and T. thermophilus (T. t) GyrB with the 43-kDa domain sequence of S. pneumoniae ParE. The residue numbering corresponds to that of the ParE sequence. The secondary structure elements deduced from the available crystal structures are shown below the sequences (boxes for helices and arrows for β sheets). Identical residues in all three sequences are highlighted in grey. ParE residues in which mutations resulted in quinolone resistance (this study) are indicated by asterisks.