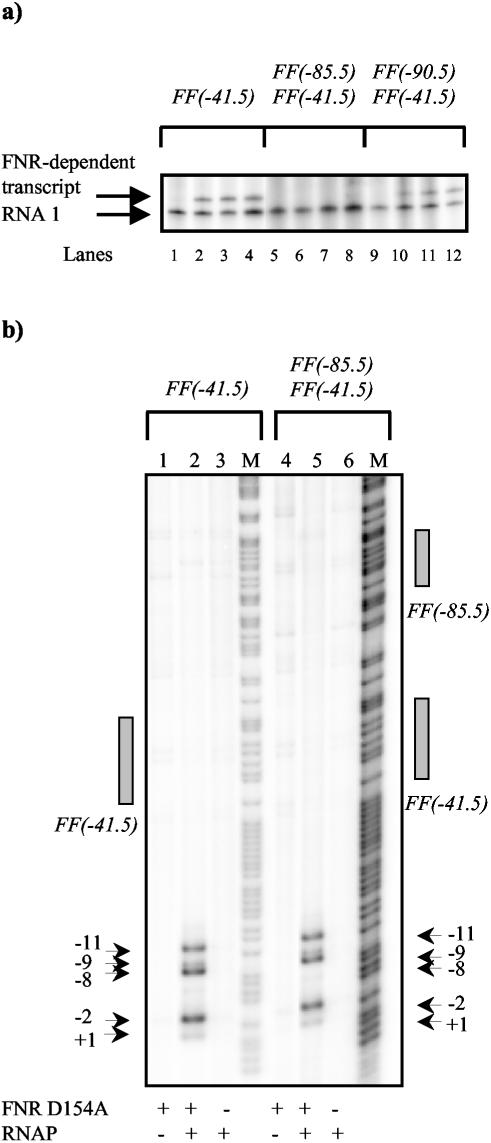
FIG. 4.
Analysis of FNR D154A-dependent open complex formation and transcription. (a) In vitro transcription analysis. The FF(−41.5)/pSR, FF(−85.5)FF(−41.5)/pSR, and FF(−90.5)FF(−41.5)/pSR constructs were used as templates for in vitro transcription. Eight nanomolar template plasmid DNA was incubated with 50 nM RNA polymerase, nucleoside triphosphates, and [α-32P]UTP, with or without FNR D154A (0 to 0.5 μM). Transcripts were analyzed on a denaturing polyacrylamide gel. The control RNA1 and the FNR-dependent transcript are marked. Samples were loaded as follows: no FNR D154A (lanes 1, 5, and 9); 0.05 μM FNR D154A (lanes 2, 6, and 10); 0.1 μM FNR D154A (lanes 3, 7, and 11); 0.5 μM FNR D154A (lanes 4, 8, and 12). (b) Permanganate footprint analysis. Shown is a phosphor image of a denaturing polyacrylamide sequencing gel on which DNA cleavage due to attack by permanganate was analyzed. [γ-32P]ATP-labeled promoter fragments were incubated with or without FNR D154A (1 μM) and with or without purified RNA polymerase σ70 holoenzyme (50 nM) prior to treatment with permanganate. The gel is calibrated with Maxam-Gilbert G+A sequencing reactions. The locations of the permanganate-induced cleavage sites and the FNR consensus binding sites are marked. Samples were loaded as follows: G+A sequencing reaction (M); 1 μM FNR D154A (lanes 1 and 4); 1 μM FNR D154A, 50 nM RNAP (lanes 2 and 5); 50 nM RNAP (lanes 3 and 6).