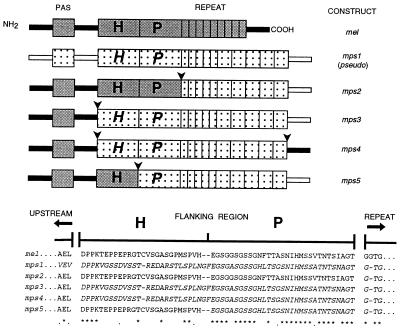
Figure 1.
(Top) per transgenes of D. melanogaster N- and C-terminal sequences (filled bars) D. pseudoobscura (open bars). The approximate positions of the N-terminal PAS domain (39) are shown as filled or speckled boxes for the two species. D. melanogaster sequences in blocks H (amino acids 639–664, ref. 2 for numbering) and P (amino acids 665–695) are in gray, and corresponding sequences in D. pseudoobscura are speckled and italicized. Thr-Gly repeats of D. melanogaster (amino acids 696–737) and D. pseudoobscura are shown as narrow repetitive units, 9 for D. melanogaster, representing the 20 pairs of Thr-Gly dipeptides, and 5 for D. pseudoobscura, representing the 10 imperfect dipeptide repeats in this species (13, 14). The 5 broader repeat units in D. pseudoobscura represent the 30 copies of the pentapeptide encoding sequence (see refs. 13 and 14). Arrows show positions of chimeric junctions. (Bottom) Coevolving amino acid blocks (marked H and P, see text). A few amino acids N-terminal to block H and the first few residues of the Thr-Gly repeat (C-terminal to block P) are shown. The length of this repeat is 66 amino acids in D. melanogaster and 209 amino acids in D. pseudoobscura (13). Asterisks indicate identical amino acids, and dots indicate conservative substitutions. D. melanogaster amino acids are shown in roman type, and D. pseudoobscura sequences are shown in italic type.