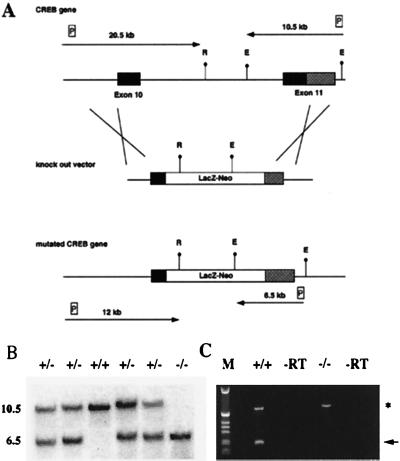
Figure 1.
Targeting strategy for the generation of a CREB null allele. (A) (Top) Gene structure of the 3′ region of the CREB locus. Exons are shown as boxes. Solid boxes, coding regions; shaded boxes, nontranslated regions. (Middle) Targeting vector. (Bottom) Gene structure of the targeted allele. E, EcoRI; R, EcoRV; LacZ-Neo, fusion gene encoding β-galactosidase and neomycin phosphotransferase; P, probes used for detection of homologous recombination events. Arrows show the expected size of the restriction fragments detected with these probes. (B) Southern blot analysis of EcoRI-digested genomic DNA from E18.5 newborns. The 3′ probe was used, the size of the restriction fragments is indicated. (C) RT–PCR analysis using a cDNA derived from total brain RNA of a CREB −/− mouse and wild-type littermate, respectively, demonstrating the absence of the 3′ part of the CREB transcript in mutant mice. Primers for the CREB gene were derived from the 3′ end of the gene; primers for Hprt (hypoxanthine guanine phosphoribosyltransferase) were used as control for RNA integrity. The CREB PCR product is indicated by an arrow, and the Hprt PCR product is indicated by an asterisk. −RT, −reverse transcriptase.