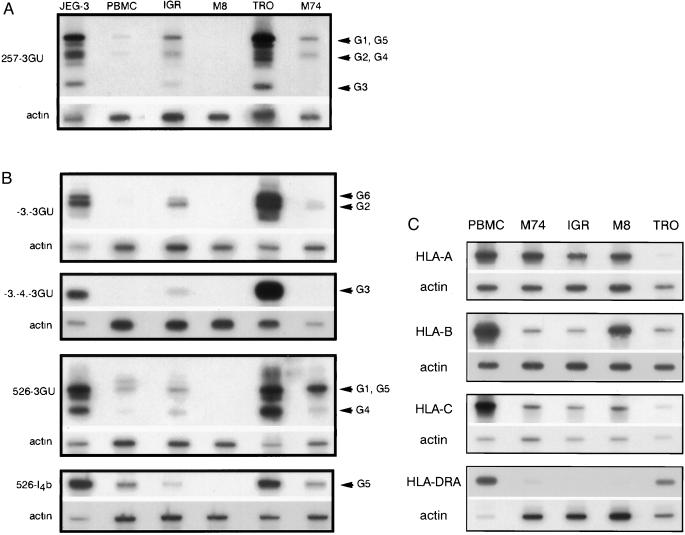
Figure 1.
(A) RT-PCR analysis of HLA-G mRNA isoforms in melanoma cells. Pan-HLA-G primers 257 (exon 2) and 3GU (3′ untranslated) were used for PCR amplification of HLA-G transcripts corresponding to all known HLA-G isoforms. cDNA from choriocarcinoma JEG-3, first trimester trophoblast (TRO), and PBMC were used as controls for high and basal HLA-G transcription. IgR, M8, and M74 correspond to cDNA amplification of melanoma cell lines. HLA-G-specific bands were revealed by hybridization with the GR-specific probe located in exon 2. Bands corresponding to HLA-G1, G2, G3, G4, and G5 transcripts are indicated by arrows. PCR products co-amplified in the same reaction by β-actin primers were detected on the same membrane by a β-actin probe. (B) Specific RT-PCR detection of alternative HLA-G transcripts in melanoma cells. Primer –3. is specific for HLA-G2 and soluble G2 (G6) isoforms that lack exon 3. Primers -3.-4. discriminate HLA-G4 mRNA transcripts. Primers 526 and I4b specifically amplify the HLA-G5 transcript, which corresponds to the soluble isoform. PCR products co-amplified in the same reaction by β-actin primers were detected on the same membrane by a β-actin probe. (C) RT-PCR analysis of classical HLA class I mRNA. cDNA from PBMC, trophoblast cells, and melanoma cell lines were amplified by RT-PCR by using locus-specific HLA-A, HLA-B, HLA-C, or HLA-DRA locus-specific primers. Locus-specific bands were revealed by using HLA-A, -B, -C, and DRA-specific probes. PCR products co-amplified in the same reaction by β-actin primers were detected on the same membrane by a β-actin probe used as a quantitative control for RNA levels.