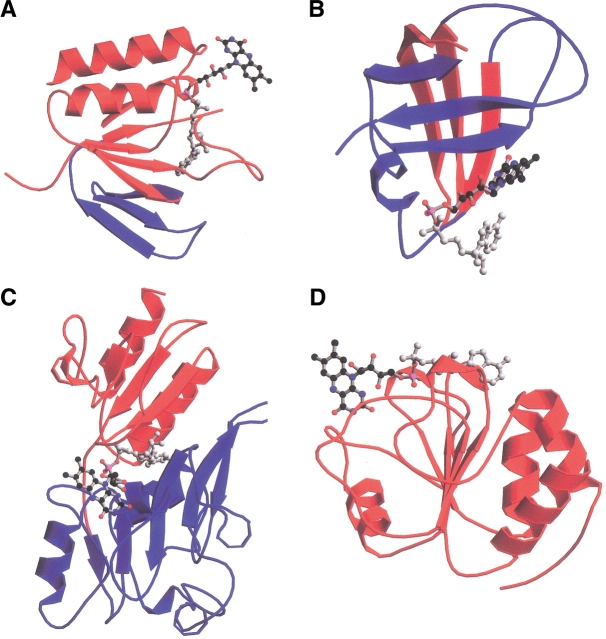
Fig. 4.
Ribbon representation of the FAD-binding domain of the four FAD-family folds complexed with FAD. (A) Rossmann fold adopted by the glutathione reductase (GR) family members. The blue shading indicates the crossover connection composed of a three-stranded antiparallel β-sheet; the red shading indicates the central five-stranded parallel β-sheet surrounded by two α-helices. The FAD cofactor adopts an elongated conformation with the adenosine moiety (gray circles) pointing toward the FAD-binding domain. (B) Ferredoxin reductase (FR) family. The two antiparallel three-stranded β-sheets are shown in red and blue. The FAD in its bent conformation is shown with the isoalloxazine ring (black circles) pointing toward the FAD-binding domain. (C) The α + β fold adopted by the p-cresol methylhydroxylase (PCMH) family members. In red are three parallel β-strands surrounded by α-helices, and in blue are five antiparallel β-strands surrounded by α-helices. The FAD molecule adopts an elongated conformation and is located in between the two subdomains with the adenine ring pointing toward them. (D) Rossmann fold adopted by pyruvate oxidase (PO) family members, shown in red. The cofactor adopts an elongated conformation and lies perpendicular to the β-strands. The stick drawing of the cofactor is depicted with gray circles for atoms in the adenine and sugar rings, with red circles for phosphate and oxygen atoms, and black circles for atoms for the isoalloxazine ring. Figure created by MOLSCRIPT (Kraulis 1991) and RASTER3D (Bacon and Anderson 1988; Merritt and Murphy 1994).