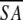The notation of the structures is as follows: <SA> are the final 30 simulated annealing structures; 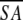 is the mean structure obtained by averaging the coordinates of the individual is the mean structure obtained by averaging the coordinates of the individual 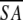 structures best fit to each other (excluding residues 1–3, 82–89, and 112–123); and ( structures best fit to each other (excluding residues 1–3, 82–89, and 112–123); and (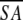 )r is the restrained minimized mean structure obtained by restrained minimization of the mean structure SA (Nilges et al. 1988a). The number of terms for the various restraints is given in parentheses. )r is the restrained minimized mean structure obtained by restrained minimization of the mean structure SA (Nilges et al. 1988a). The number of terms for the various restraints is given in parentheses. |
|
a For backbone NH-CO hydrogen bond there are two restraints: rNH-O = 1.5–2.3 Å and rN-O = 2.5–3.3 Å. All hydrogen bonds involve slowly exchanging NH protons. |
|
b The torsion angle restraints comprise 110 φ and 110 ψ. |
|
c The values of the square-well NOE (FNOE) and torsion angle (Ftor) potentials (cf. Equations 2 and 3 in Clore et al. 1986) are calculated with force constants of 50 kcal mol−1 Å −2 and 200 kcal mol−1 rad−2, respectively. |
|
d The value of the quartic van der Waals repulsion term (Frep) (cf. Equation 5 in Nilges et al. 1988c) is calculated with a force constant of 4 kcal mol−1 Å −4 with the hard-sphere van der Waals radius set to 0.8 times the standard values used in the CHARMM (Brooks et al. 1983) empirical energy function (Nilges et al. 1988a,b,c). |
|
e EL-J is the Lennard-Jones-van der Waals energy calculated with the CHARMM empirical energy function and is not included in the target function for simulated annealing or restrained minimization. |
|
f The improper torsion restraints serve to maintain planarity and chirality. |
|
g These were calculated by using the PROCHECK program. |
|
h The residues in the regular secondary structure are 17–36(α1), 39–45(α2), 58–72(α3), 101–108(α4), 4–10(β1), 49–54(β2), 75–78(β3) and 92–98(β4). |
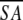 )r
)r
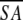 is the mean structure obtained by averaging the coordinates of the individual
is the mean structure obtained by averaging the coordinates of the individual 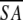 structures best fit to each other (excluding residues 1–3, 82–89, and 112–123); and (
structures best fit to each other (excluding residues 1–3, 82–89, and 112–123); and (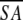 )r is the restrained minimized mean structure obtained by restrained minimization of the mean structure SA (Nilges et al. 1988a). The number of terms for the various restraints is given in parentheses.
)r is the restrained minimized mean structure obtained by restrained minimization of the mean structure SA (Nilges et al. 1988a). The number of terms for the various restraints is given in parentheses.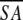 )
)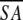 )r
)r