Figure 1.
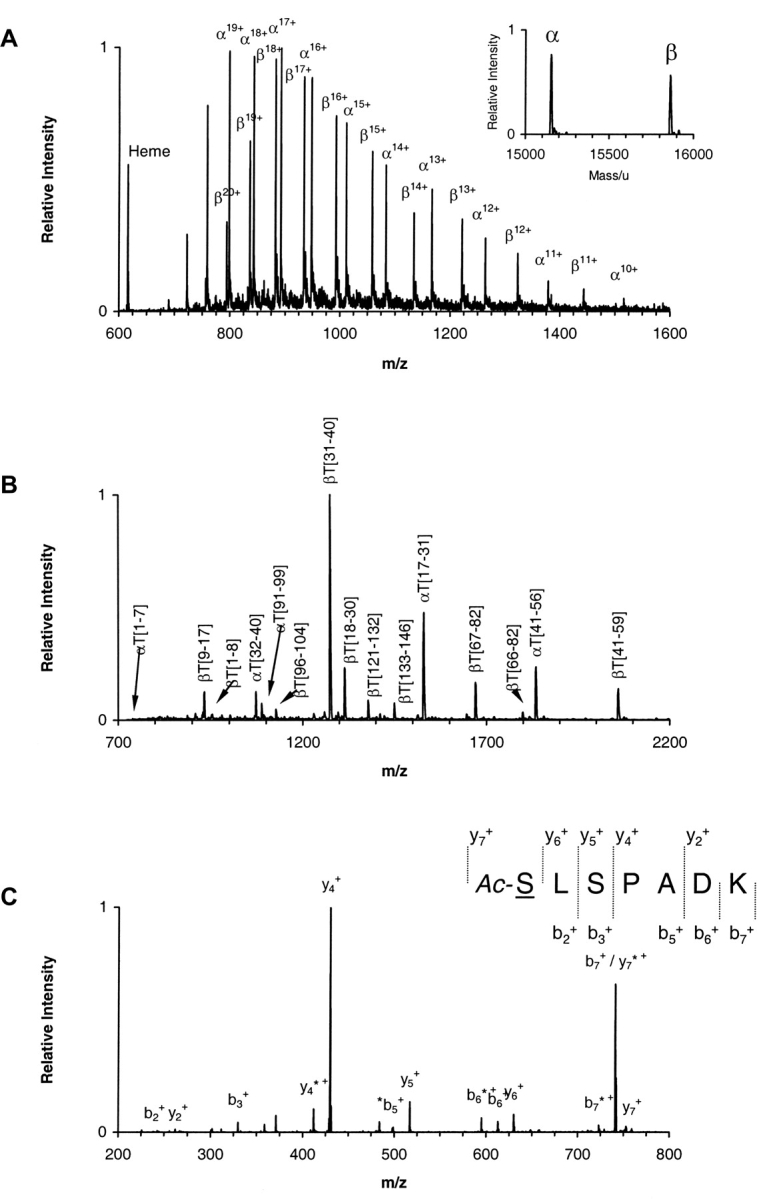
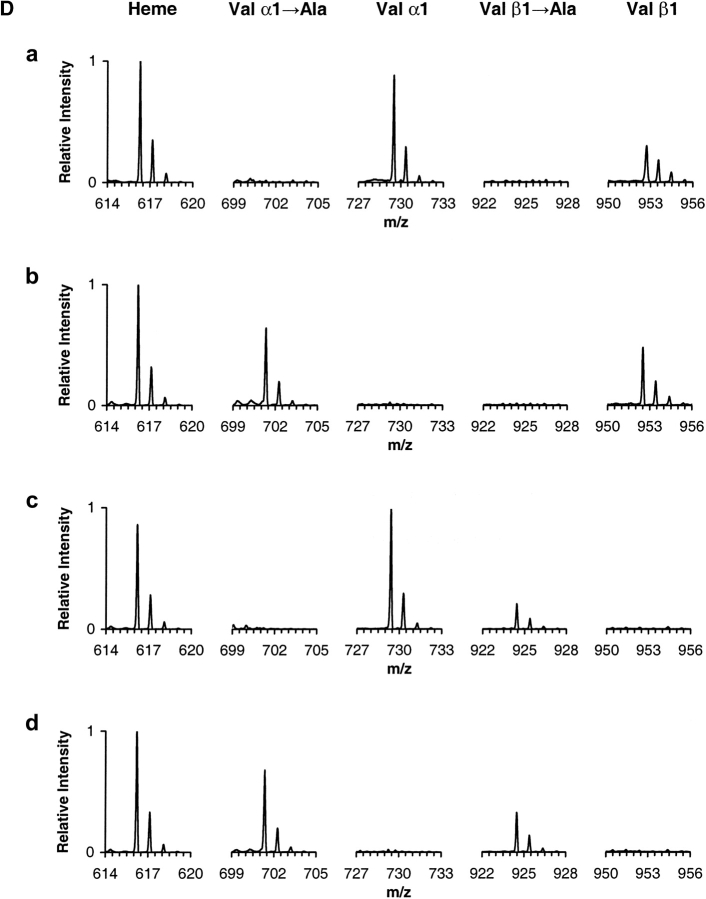
Mass spectrometry analysis. (A) ESI/MS analysis of hemoglobin sample α2AcSβ2. The ion envelopes for α and β chains are shown in the raw m/z spectrum. (Inset) Transformed mass spectrum after deconvolution of the ion envelopes encompassing the region of charge states 11+ through 20+ (m/z range 750–1500). (Refer to text for experimental details.) (B) MALDI-QqTOF/MS spectrum of a tryptic digest of hemoglobin sample α2AcSβ2. Peaks are labeled as α- or β-chain peptides followed by T for trypsin and the number of the first and last residues in the peptide in accordance with the numbering system for the primary structure of the entire polypeptide chain. The combined MALDI data yielded information covering 100% of both α and β globin chains. (C) ESI/MS/MS analysis of the singly charged peak of the α-chain N-terminal tryptic peptide αT(1–7) from hemoglobin sample α2AcSβ2. The underlined residue was mutated. Only b and y ions are labeled for clarity. Unidentified peaks are labeled with an asterisk. The mutated residue was assigned based on the occurrence of ions y6+ and y7+ and the PTC-derivative of the same peptide. (Refer to text for experimental details.) (D) ESI/MS/MS hypothesis-driven analysis of the N-terminal tryptic peptide αT(1–7) of human hemoglobin (row a) and recombinant hemoglobin samples α2Aβ2 (row b), α2β2A (row c) and α2Aβ2A (row d). Spectra show the isolated precursor ions selected based on either the presence or absence of the Val → Ala modification. The hypothetical precursor ions were subsequently fragmented.