Figure 3.
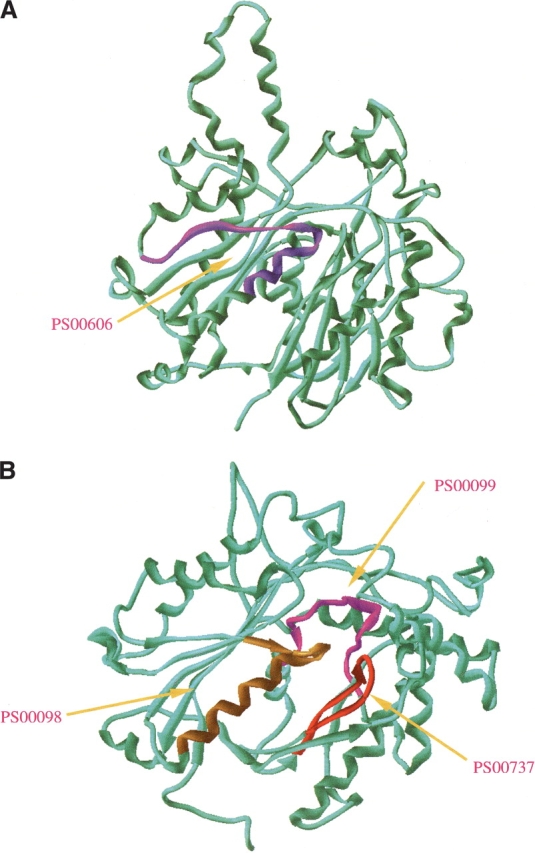
Ribbon representation of two remote homologs (1dd8A and 1afwA), confidently recognized by using the PROSITE motif-based descriptors. (A) 3D model of 1dd8A (β-ketoacyl [acyl carrier protein] synthase I from Escherichia coli; Chain: A; FSSP family index: 103.1.1.1.1.1) (Olsen et al. 1999); (B) 3D model of 1afwA (thiolase from Saccharomyces cerevisiae; Chain: A; FSSP family index: 103.1.1.1.2.1) (Mathieu et al. 1997). Although the sequence identity for the two proteins was only 12%, they were found to share high structural similarity by using CE (the RMSD for 229 structural aligned residues is 3.1 Å and the Z-score is 5.0). Taking the 1dd8A as the query sequence against the other proteins in FSSP607, the retrieved top hit was 1afwA with a score (i.e., MOTIF_SCOP_SIM) of 8.599. The PROSITE motifs contributing in this recognition were PS00606 (entry name: B_KETOACYL_SYNTHASE; pattern: G-x(4)-[LIVMFTAP]-x(2)-[AGC]-C-[STA](2)-[STAG]-x(3)-[LIVMF]), PS00098 (entry name: THIOLASE_1; pattern: [LIVM]-[NST]-x(2)-C-[SAGLI]-[ST]-[SAG]-[LIVMFYNS]-x-[STAG]-[LIVM]-x(6)-[LIVM]), PS00737 (entry name: THIOLASE_2; pattern: N-x(2)-G-G-x-[LIVM]-[SA]-x-G-H-P-x-[GA]-x-[ST]-G), and PS00099 (entry name: THIOLASE_3; pattern: [AG]-[LIVMA]-[STAGCLIVM]-[STAG]-[LIVMA]-C-x-[AG]-x-[AG]-x-[AG]-x-[SAG]). The localization of these motifs in the 3D models was marked with different colors in the two proteins. Interestingly enough, a remote homology was recognized by the MOTIF_SCOP_SIM similarity function due to the high SFM scores for these motifs within the SCOP tillage-like fold, although none of the motifs was found to co-occur in the two proteins.
