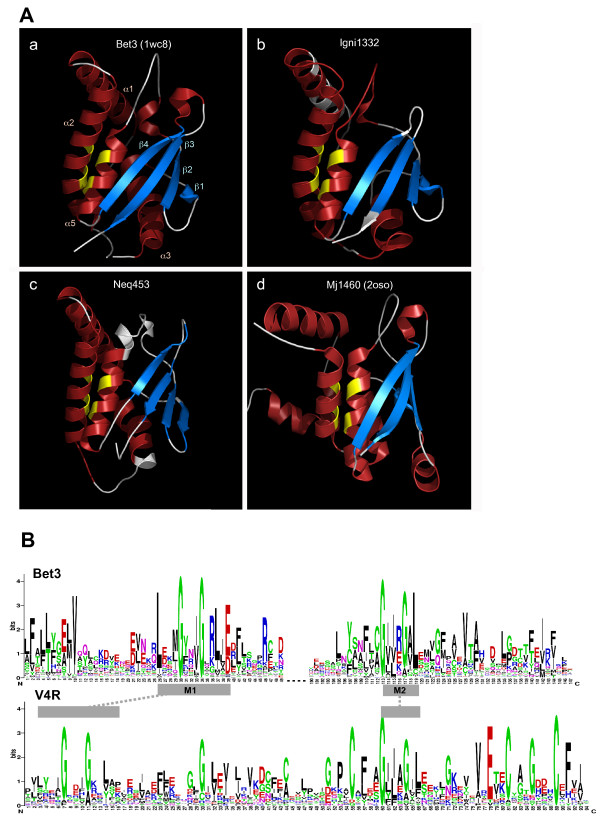
Figure 1.
(A) Structure of mouse BET3 (PDB:1wc8)(a) and homology models of archaeal V4R domains (Ignicoccus hospitalis p1332 and Nanoarchaeum equitans p453)(b, c). Secondary structure inference for the archaeal proteins is overlaid on the models, coloring of the alpha helix (red) and beta strand (blue) elements corresponding to the type of prediction using PSIPRED v2.5. The location of the conserved pairs of glycines in helices 2 and 5 is indicated in yellow. The crystal structure of the Methanocaldococcus jannaschii p1460 (PDB, 2oso) is shown in panel d. The models were visualized using Pymol [17]. (B) Sequence conservation logos for eukaryotic TRAPP-Bet3 and archaeal V4R domain proteins. M1 and M2 represent previously identified motifs characteristic to the TRAPP-Bet3 family and are linked to the corresponding structure and sequence regions in V4R. The sequence logos were generated using WebLogo3 [18](the sequence alignments used as input are available upon request).