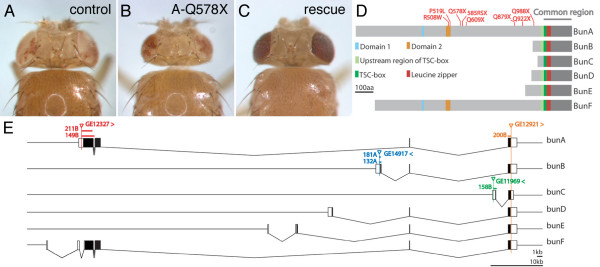
Figure 1.
Identification of bunA as positive growth regulator. Eye-specific reduction of bunA function by means of eyFLP/FRT-mediated mitotic recombination results in a reduction of eye and head size (B) as compared to the control (A). This growth deficit is rescued by overexpression of a bunA transgene (C). (D) Schematic representation of the six Bun protein isoforms. The putative Bun transcription factors have distinct N-termini but an identical C-terminal region (common region), including the TSC-box (DNA-binding) and an adjacent leucine zipper (homo- and heterodimerization) encoded by the very 3' bun exon (E). All Bun isoforms except for BunC also contain a conserved region N-terminally to the TSC-box that is present in all mammalian TSC-22/Dip/Bun family members. In addition, BunA and BunF possess two domains in their N-terminal regions that are conserved among mammalian homologs ([13], domain 1 aligns to BunA amino acids 369-82 [Swissprot:Q24523-1]). The eight EMS-induced mutations isolated in the eyFLP/FRT screen (indicated in red) affect only BunA and BunF. (E) The genomic region of bun according to FlyBase [39]. The six bun transcripts share the last exon but have distinct 5' exons. UTRs are shown in white and ORFs in black. P-element insertions used for the jump-out screens and deletions obtained in these screens are indicated. Arrowheads indicate the directions of transcription that can be driven by the respective EP insertions. The P-element GE12327 and the deletions derived from it as well as the EMS-induced alleles affect both bunA and bunF but are referred to as bunA alleles. Genotypes are: (A) y, w, eyFLP/y, w; FRT40A, w+, cl2L3/FRT40Aiso; (B) y, w, eyFLP/y, w; FRT40A, w+, cl2L3/FRT40A, bunA-Q578X; (C) y, w, eyFLP/y, w; FRT40A, w+, cl2L3/FRT40A, bunA-Q578X; ey-Gal4, GMR-Gal4/UAS-bunA.