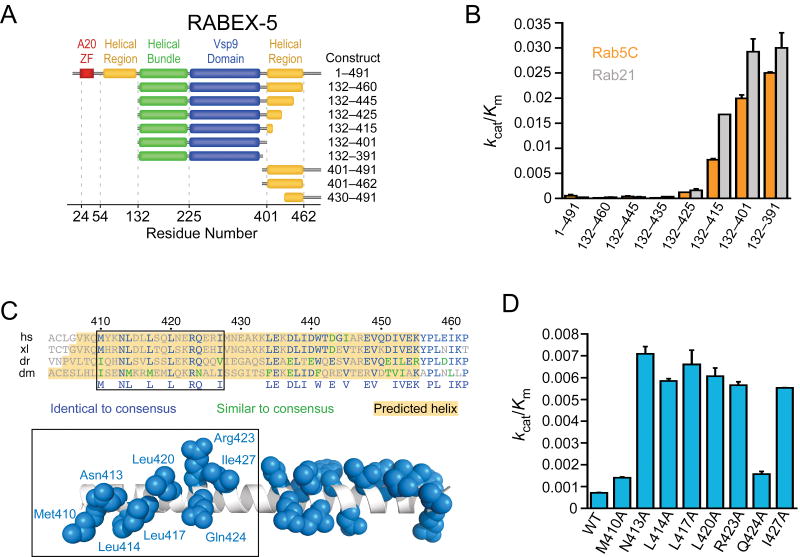
Figure 4.
Identification of an autoinhibitory element in RABEX-5. (a) RABEX-5 truncation constructs. (b) Catalytic efficiency of RABEX-5 truncation constructs. Values are mean ± s.d. for n = 2. (c) Alignment of RABEX-5 sequences from diverse organisms (Hs, Homo sapiens; Xl, Xenopus laevis; Dr, Danio rerio; Dm, Drosophila melanogaster). Below the alignment is a hypothetical helical model for RABEX-5407–456 with conserved side chains depicted in blue (in the default rotamer conformation). The helix was constructed using PyMOL. (d) Effect of alanine substitutions on the catalytic efficiency of RABEX-5132–460. Values are mean ± s.d. for n = 2.