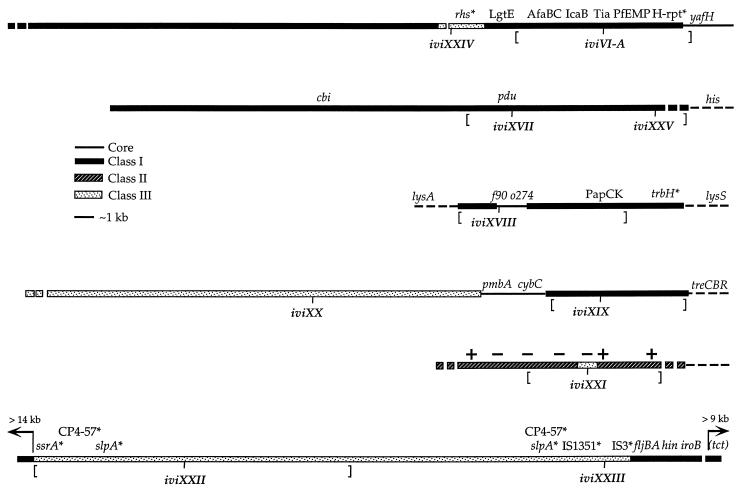
Figure 1.
Salmonella-specific regions identified by IVET. Each line represents an assemblage of overlapping λ and/or plasmid clones in a Salmonella-specific region. The boxed sequences represent Salmonella-specific regions that are distinct to Salmonella spp. and are not present in other enteric species tested. Class I sequences hybridized to all Salmonella serovars. Class II sequences hybridized to all Salmonella serovars tested with the exception of S. typhi. Class III sequences hybridized only to broad host-range serovars (S. typhimurium and S. newport). Core sequences are those shared between Salmonella spp. and E. coli. Each region was identified by hybridization to a probe from an ivi gene fragment of atypical base composition as determined by sequencing >200 bp starting from the ivi junction. Protein designations indicate predicted similarities ranging from 30–81% identity at the amino acid level over a minimum of 50 deduced residues. Protein similarities include enterotoxigenic E. coli (ETEC) Tia (10, 12) and AfaBC, involved in afimbrial adhesin in uropathogenic and diarrheal-associated E. coli strains (13), PfEMP-1, involved in attachment of Plasmodium falciparum-infected red blood cells to peripheral epithelium (10, 14) and IcaB, an intercellular adhesin molecule of Staphylococcus epidermidis that functions in biofilm formation (15). LgtE of Neisseria meningitidis is involved in modification of surface lipopolysaccharide (16). PapC and PapK are adherence-related functions of uropathogenic E. coli (UPEC) (17, 18). Gene designations denote similarity ranging from 61–96% at the DNA level over a minimum of 129 nucleotides. The S. typhimurium open reading frames f90 and o274 are homologues of E. coli genes that do not map to the corresponding region of E. coli (minute 47 in E. coli vs. minute 65 in S. typhimurium). Similarly, Salmonella pap-like sequences map to minute 65, which is near, but distinct from, the corresponding papCK-containing region of uropathogenic E. coli at minute 67 (19). Brackets designate the endpoints of deletions tested in the CI assay. The “+” designation over the iviXXI region indicates that the corresponding chromosomal segment hybridizes to DNA prepared from the laboratory strain of S. typhimurium, LT2; a “−” designation indicates that it does not hybridize to DNA prepared from LT2. For all other regions, probes hybridized equally well to DNA prepared from S. typhimurium strains 14028 and LT2. The ∗ designates homologies that are associated with mobile genetic elements that are discussed in the text with the exception of trbH, which is associated with a pathogenicity island of Dichelobacter nodosus (20).