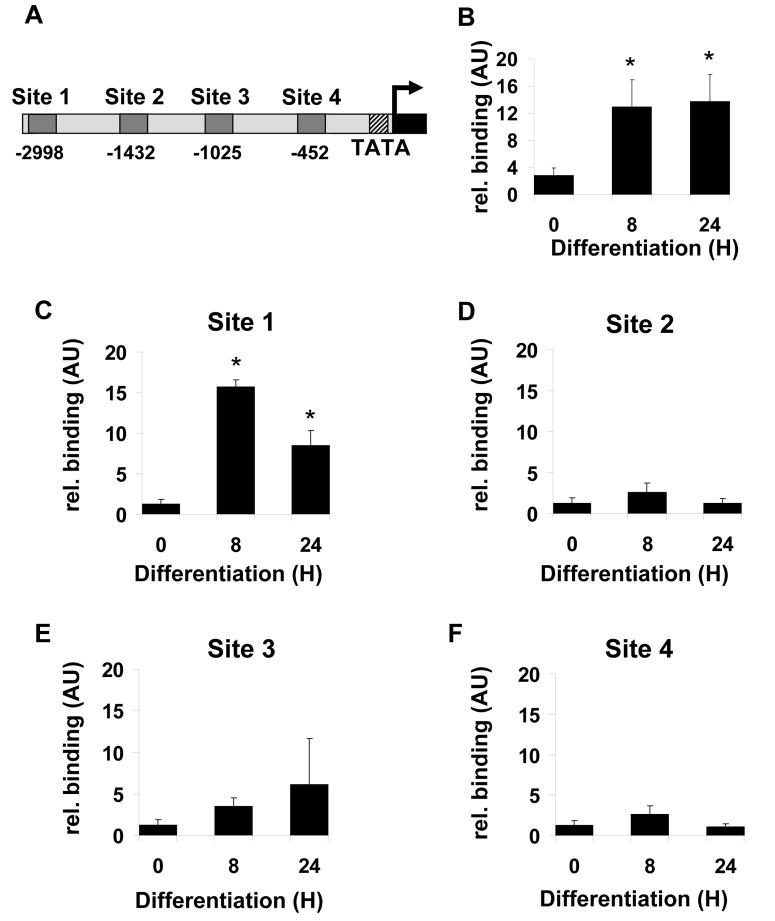
Figure 6. Identification of C/EBPβ binding sites in the DGAT2 promoter.
A, analysis of 3kb upstream of the first exon of DGAT2 by TESS revealed four potential C/EBPβ binding sites, denoted site 1 to site 4. 3T3-L1 preadiocytes were induced to differentiate for various times and subjectd to ChIP analysis to assess binding of C/EBPβ to the well characterised site in the C/EBPα promoter (B) or the putative site 1 (C), site 2 (D), site 3 (E) or site 4 (F) in the DGAT2 promoter. C/EBPβ bound DNA in immunoprecipitates was quantified by real-time PCR. Values in each sample were normalised to total genomic DNA of the same sequence in the “input” starting sample prior to immunoprecipitation. Data are the average of 6 independent experiments +/− SEM, * indicates statistically significant difference from binding at time 0 (p<0.05).