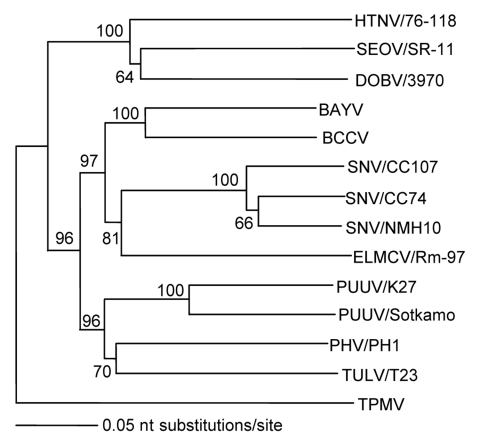
Figure 2.
Phylogenetic relationship between Thottapalayam virus (TPMV) and other hantaviruses based on the nucleotide sequences of the full-length small (S) genomic segment, determined by using the neighbor-joining method. Numbers at each node are bootstrap probabilities (expressed as percentages) determined for 1,000 iterations. Branch lengths are proportional to number of nucleotide substitutions per site. Sequences used for comparison were those of Hantaan (HTNV/76–118, NC 005218), Seoul (SEOV/SR-11, M34881), Dobrava (DOBV/3970, L41916), Bayou (BAYV, L36929), Black Creek Canal (BCCV, L39949), Sin Nombre (SNV/CC107, L33683; SNV/CC74, L33816; and SNV/NMH10, L25784), El Moro Canyon (ELMCV/Rm-97, U11427), Puumala (PUUV/K27, L08804 and PUUV/Sotkamo, NC 005224), Prospect Hill (PHV/PH1, Z49098), and Tula (TULV/T23, Z30945) viruses. Strain designations are unavailable for BAYV and BCCV. The full-length S-segment sequence of TPMV has been deposited into GenBank (accession no. AY526097).