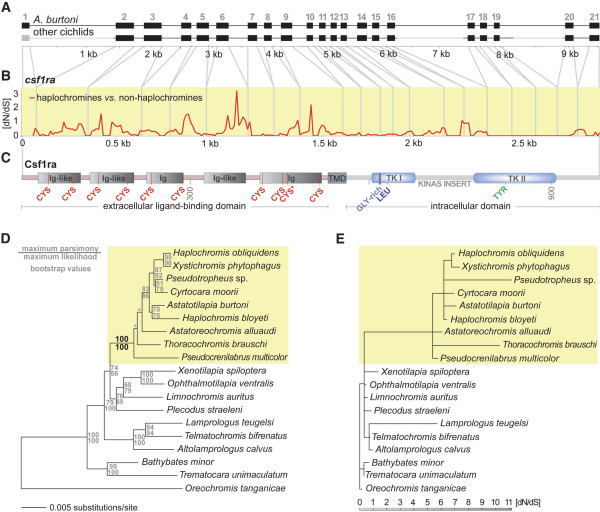
Figure 4.
Molecular evolutionary analyses of the csf1ra locus. (a) The entire csf1ra gene locus was obtained from 19 cichlid species (exon 1 and the relatively large introns 1, 16, and 19, which are shown in gray, were not sequenced). (b) dN/dS ratio of haplochromines compared to non-haplochromine cichlids as revealed from a sliding window analysis with DNASP. (c) Schematic representation of the structure of the Csf1ra protein. The gene consists of an extracellular ligand-binding domain containing five immunoglobulin-like (Ig-like) domains with conserved cysteines (CYS), a transmembrane domain (TMD), and an intracellular partition that contains two tyrosine kinase (TK) domains interrupted by a kinase insert domain. In the tyrosine kinase I domain (TK I) a glycine-rich region (GLY-rich) and a conserved leucine (LEU) is found, the tyrosine kinase II domain (TK II) contains a conserved tyrosine (TYR). The asterisk indicates that in one species, X. phytophagus, the otherwise conserved cysteine at amino acid position 450 has been replaced by a tryptophan. (d) Maximum likelihood phylogeny based on more than 4100 bp of non-coding sequences of the csf1ra gene locus corroborating the monophyly of the haplochromines (yellow box) and the ancestral position of A. alluaudi, P. multicolor, and T. brauschi [7]. The asterisk indicates relatively short branches not supported by high bootstrap values or a Shimodaira-Hasegawa test. (e) Branch-scaled tree showing the dN/dS rates reconstructed with HyPhy. The only internal branch with a dN/dS > 1 is the one representing the common ancestor of the haplochromines (yellow box).