Figure 1a.
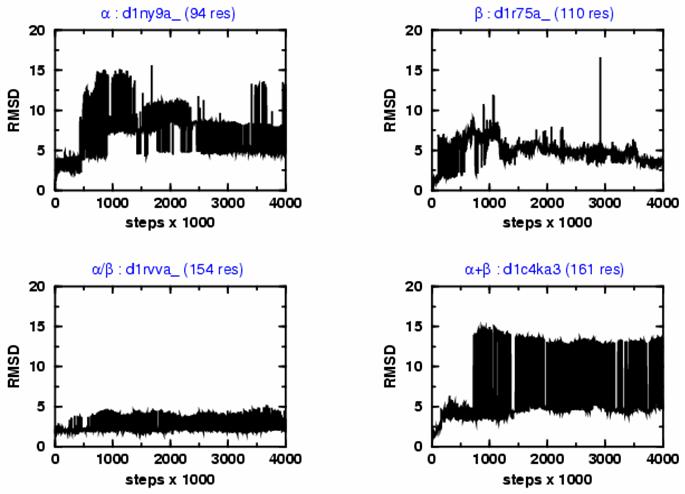
Showing the variation of the root mean square deviation of all Cα atoms (RMSD) from the native structure with the number of Monte Carlo iteration steps for four SCOP protein domains representing the four major structure classes: α (d1ny9a_), β (d1r75a_), α/β (d1rvva_), and α+β (d1c4ka3). Domains are described with the 3-point per residue model. All trajectories started from the crystal structures and were propagated for a total of 4,000,000 steps using an advanced combination of Parallel Tempering and Equi-Energy Monte Carlo methods. The simulation explore the conformational space around the native structure with rapid and frequent transitions between states that have very different RMSD values; at the end of the trajectories, the RMSD generally reaches values lower than 5 Å.
