Abstract
Cys synthesis in plants takes place in plastids, cytosol, and mitochondria. Why Cys synthesis is required in all compartments with autonomous protein biosynthesis and whether Cys is exchanged between them has remained enigmatic. This question was addressed using Arabidopsis thaliana T-DNA insertion lines deficient in the final step of Cys biosynthesis catalyzed by the enzyme O-acetylserine(thiol)lyase (OAS-TL). Null alleles of oastlA or oastlB alone showed that cytosolic OAS-TL A and plastid OAS-TL B were completely dispensable, although together they contributed 95% of total OAS-TL activity. An oastlAB double mutant, relying solely on mitochondrial OAS-TL C for Cys synthesis, showed 25% growth retardation. Although OAS-TL C alone was sufficient for full development, oastlC plants also showed retarded growth. Targeted affinity purification identified the major OAS-TL–like proteins. Two-dimensional gel electrophoresis and mass spectrometry showed no compensatory changes of OAS-TL isoforms in the four mutants. Steady state concentrations of Cys and glutathione and pulse-chase labeling with [35S]sulfate indicated strong perturbation of primary sulfur metabolism. These data demonstrate that Cys and also sulfide must be sufficiently exchangeable between cytosol and organelles. Despite partial redundancy, the mitochondria and not the plastids play the most important role for Cys synthesis in Arabidopsis.
INTRODUCTION
Cys is a proteinogenic amino acid and thus indispensable for any living cell. It must be provided from outside or synthesized inside the cell. Animal cells either take up Cys with their diet or synthesize it from Met via transsulfurylation of cystathionine in the cytosol. Yeast cells can take up external Cys and are also able to synthesize Cys in the cytosol from sulfide and O-acetylhomoserine and by transsulfurylation (Thomas et al., 1997). Mitochondria in yeast and animal cells must receive Cys for their protein translation machinery from the cytosol, although little is known about such amino acid permeases (Düring-Olsen et al., 1999; Reinders et al., 2006). In plants, the situation is entirely different. Not only are three compartments with independent protein biosynthesis present in the cell, but Cys can be synthesized in each of these compartments: plastids, mitochondria, and the cytosol. Why Cys biosynthesis is organized in this way remains unexplained to date; nor is anything known about special or redundant functions or communication between the three cellular locations. The biosynthesis of Cys is a tripartite process, which can be subdivided into (1) synthesis of the carbon- and nitrogen-containing precursor of Cys, O-acetylserine (OAS), (2) generation of sulfide by assimilatory reduction of sulfate, and (3) combination of OAS and sulfide to produce Cys. Serine acetyltransferase (SAT; EC 2.3.1.30) catalyzes the formation of OAS by transferring an acetyl moiety from acetyl-CoA to Ser. Subsequently, O-acetylserine(thiol)lyase (OAS-TL; EC 2.5.1.47) replaces the activated acetyl moiety in OAS with sulfide to release Cys. The exclusive location of assimilatory sulfate reduction in plastids suggests that sulfide is able to reach Cys synthesis in cytosol and mitochondria, possibly by diffusion of H2S through membranes (Jacques, 1936).
Lunn et al. (1990) suggested that endomembranes may be impermeable to Cys transport due to the reactivity of the thiol group, pointing out that each compartment with the capacity for protein biosynthesis would require its own Cys production. Alternatively, cytosolic Cys synthesis might function as a scavenging reaction for free sulfide to prevent deleterious interactions. Similarly, the mitochondrial respiratory chain might need protection against the reactivity of sulfide, while a steady supply of reduced sulfur from Cys is required for numerous essential pathways in mitochondria, such as biosynthesis of iron/sulfur clusters, thiamin, lipoate, and biotin (Droux, 2004). The exchange of sulfur-related metabolites between cytosol and plastids is also a prerequisite for the coordination of Met and S-adenosylmethionine homeostasis and the synthesis of GSH (reviewed in Wirtz and Droux, 2005). The contradictory distribution of SAT and OAS-TL activities adds to the unknown functions of the subcellular compartmentation. The major part of OAS-TL activity in leaves of Datura innoxia is located in the cytoplasm (∼45%) and the plastids (∼45%), while only a minor part (10%) is associated with the mitochondrial fraction (Kuske et al., 1996). This pattern of distribution is confirmed by subcellular fractionation of leaves from spinach (Spinacia oleracea) (Lunn et al., 1990) and pea (Pisum sativum) (Droux, 2003). By contrast, the major part of SAT activity appears to reside in mitochondria (76 to 88%), while only residual amounts of SAT activity are found in cytoplasm (6 to 14%) and plastids (6 to 10%) from leaves of pea (Ruffet et al., 1995). This means that OAS-TL:SAT activity ratios are 200:1 in cytosol and 300:1 in plastids but only 4:1 in mitochondria, indicating that SAT is the rate-limiting step in Cys synthesis. Earlier studies found 35% of SAT activity in chloroplasts of spinach (Brunold and Suter, 1982) and 30% in mitochondria from bean (Phaseolus vulgaris) leaves (Smith, 1972), without any further attribution of the remaining SAT activities. The localization studies for SAT and OAS-TL were performed using distant taxa and therefore could reflect differences in sulfur metabolism between plant species. For that reason, a functional explanation cannot be derived from these reported distributions of SAT and OAS-TL activities per se.
OAS-TL–like proteins are formally β-substituted Ala synthases and belong to the superfamily of pyridoxalphosphate-containing enzymes (Hatzfeld et al., 2000). In Arabidopsis thaliana, SAT and OAS-TL proteins are encoded by small gene families and distributed between cytoplasm, mitochondria, and plastids (Hell et al., 1994; Bogdanova et al., 1995; reviewed in Hell et al., 2002; Kawashima et al., 2005). According to the Genevestigator database (Zimmermann et al., 2004), the most strongly transcribed genes, out of nine OASTL-like genes, are OASTLA1 (At4g14880), OASTLB (At2g43750), OASTLC (At3g59760), and CYSC1 (At3g61440). OASTLA2 produces no functional protein due to an in-frame stop codon and an unspliced intron (Jost et al., 2000); hence, the OASTLA1 gene is termed OASTLA here and encodes the only OAS-TL A–type protein in Arabidopsis. OAS-TL A (cytosolic), OAS-TL B (plastid), and OAS-TL C (mitochondrial) catalyze true OAS-TL reactions leading to Cys (Wirtz et al., 2004). By contrast, CysC1 encodes a mitochondria-localized β-cyano-Ala synthase (CAS; EC 4.4.1.9) that uses Cys to catalyze the detoxification of cyanide in a partial backward reaction of the OAS-TL type (Hatzfeld et al., 2000; Warrilow and Hawkesford, 2000; Yamaguchi et al., 2000). The remaining OASTL-like genes (CysD1 [At3g04940], CysD2 [At5g28020], CS26 [At3g03630], and CS-like [At5g28030]) are less strongly expressed, and the encoded proteins are little characterized or not at all (Hatzfeld et al., 2000; Yamaguchi et al., 2000).
The position of Cys biosynthesis between the assimilation of inorganic sulfate and the metabolism of organic sulfide makes it a prime target for the coordination of both processes. SAT and OAS-TL isoforms are not only present in the same compartments but form the decameric Cys synthase complex (CSC). Several lines of evidence suggest a dual function of the CSC in sensing the sulfur status and regulating SAT activity in response to the actual sulfur supply (Droux et al., 1998; Hell and Hillebrand, 2001; Wirtz and Hell, 2006). Unexpectedly, channeling of the intermediate OAS is not a function of complex formation, since the second enzyme OAS-TL is substantially inactivated by interaction with SAT in plants and bacteria (Kredich et al., 1969; Droux et al., 1998; Wirtz et al., 2001). Instead, OAS has been shown to dissociate the CSC in vitro, while sulfide stabilizes the interaction. Furthermore, SAT in the absence of OAS-TL is less active, and a 100-fold excess of OAS-TL protein is required to achieve maximal formation of Cys from Ser, acetyl-CoA, and sulfide (Droux et al., 1998; Mino et al., 2001). Thus, the association with OAS-TL promotes the activity of SAT. Reversible protein–protein interaction driven by the presence of OAS (Berkowitz et al., 2002) and sulfide (Kredich et al., 1969; Wirtz and Hell, 2006) thus couples Cys synthesis to the sulfur status: if sulfate becomes scarce, sulfide is missing to stabilize the CSC, and OAS, accumulated from SAT activity, dissociates the complex. This results in lowered SAT activity, reduced acetyl-CoA consumption, and OAS formation until the cell is resupplied with sulfate. Biochemical studies suggest that the CSC operates similarly in cytosol, mitochondria, and plastids (reviewed in Wirtz and Hell, 2006). The combination of CSC properties with the uneven distribution of SAT and OAS-TL activities in the compartments, at least of pea (Ruffet et al., 1995; Droux, 2003), suggests that Cys synthesis in mitochondria will hardly reach maximal rates due to the low OAS-TL:SAT activity ratio. The intermediate OAS may actually leave mitochondria to supply Cys synthesis for the cytosol or even plastids. By contrast, the high OAS-TL:SAT ratio in plastids will prevent any loss of OAS to the cytosol. When the equilibrium between free and CSC-associated SAT was disrupted in the cytosol of tobacco (Nicotiana tabacum) plants, Cys synthesis was deregulated between the other compartments, confirming the pace-controlling function of the CSC in vivo (Wirtz and Hell, 2007).
The cellular coordination of Cys synthesis was investigated in Arabidopsis by determination of the levels and impact of OAS-TL activities in different compartments. T-DNA knockout lines of the major OAS-TL isoforms A (cytoplasm), B (plastids), and C (mitochondria) in combination with SAT-based affinity purification of the OAS-TL protein family showed that OAS-TL A and B dominated by far in abundance and activity. However, their absence caused no visible phenotype. By contrast, the seemingly small contribution of mitochondrial OAS-TL C activity was sufficient for the survival of an oastlAB double mutant; its absence caused growth retardation of an oastlC null mutant. Thiol contents and tracer studies with 35S showed that the phenotypes of oastlAB and oastlC were based on different mechanisms. These data demonstrate that Cys and sulfide must be exchanged between the organelles and the cytosol, that the compartments can compensate for Cys deficiency to some extent among themselves, and that mitochondrial OAS-TL C is essential for normal growth.
RESULTS
Genomic Characterization of oastlA, oastlB, and oastlC Insertion Lines
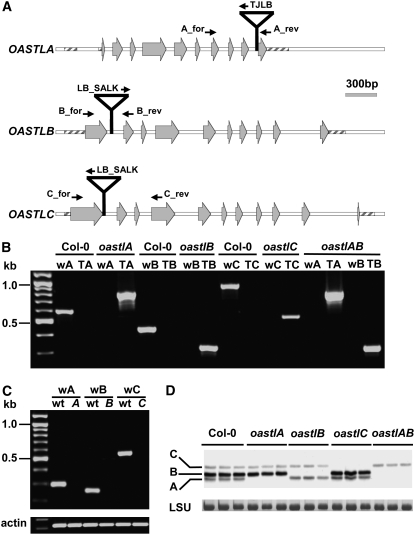
T-DNA insertion lines were selected to identify loss-of-function mutants for the genes encoding cytosolic OAS-TL A, plastid OAS-TL B, and mitochondrial OAS-TL C proteins. For the genomic characterization and selection of homozygous plants, a PCR-based screening strategy was set up using gene-specific primers to bind upstream and downstream of the predicted T-DNA insertions in combination with a primer for the left border region of the corresponding T-DNA (Figure 1A). Sequencing of the genomic regions flanking the T-DNA on both sides defined the insertions in the first intron of oastlB and oastlC at positions +248 bp and +358 bp, respectively, near the predicted sites. The T-DNA in the oastlA mutant was located in the ninth intron at +1746 bp downstream of the ATG initiation site. Homozygous T-DNA insertion lines were selected by segregation analyses as well as by PCR (Figure 1B). The presence of only a single T-DNA insertion locus in each oastl T-DNA line was independently confirmed by DNA gel blotting (see Supplemental Figure 1 online). Based on these findings, lines oastlA and oastlB were crossed and a homozygous double insertion line was identified by PCR. The resulting double mutant was termed oastlAB.
Figure 1.
Genomic Organization of oastl T-DNA Insertion Lines.
(A) Structures of genes OASTLA, OASTLB, and OASTLC are shown with insertion sites of T-DNAs in knockout plant lines. Exons are indicated by gray arrows, primers for the genetic characterization by small black arrows, and untranslated regions by striped boxes.
(B) Genomic characterization of T-DNA insertion lines. PCR with genomic DNA as template showed the presence of T-DNA in each oastl gene in the appropriate mutant; lack of the wild-type–specific product showed that the T-DNA allele was homozygous. Each wild type (w) and T-DNA (T) primer pair was specific for the gene of the indicated mutant.
(C) T-DNA insertions resulted in loss of mRNA of the three oastl genes as shown by RT-PCR. Amplification of actin from the same cDNA preparations was used as a positive control.
(D) Immunoblot of leaf protein extracts from the indicated oastl insertion lines (three lanes each) detected with a polyclonal antibody against Arabidopsis OAS-TL C. Cross-reaction with OAS-TL isoforms A and B shows the specific knockout of the indicated isoforms of OAS-TL in the mutants. Staining intensities of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (LSU) protein in the same samples confirm equal loading in the individual lanes.
The expression of OASTL genes was analyzed by RT-PCR using the gene-specific primers described before. In none of the three single mutants was the corresponding transcript detected (Figure 1C), indicating complete gene knockouts. In the oastlA mutant, no truncated transcripts in front of the insertion in the ninth intron could be detected (data not shown). The four oastl lines were tested for the presence of OAS-TL A, B, and C in leaf protein extracts using an anti-At OAS-TL C polyclonal antiserum that cross-reacts with OAS-TL A and B (Jost et al., 2000). From bottom to top, three bands were detected in wild-type plants that corresponded to OAS-TL A, B, and C (Figure 1D). The immunoblot demonstrated that the T-DNA insertions in the oastlA, oastlB, and oastlC genes prevented any detectable synthesis of the encoded proteins. Strict correlations of band intensities with relative abundance of the proteins were not expected: OAS-TL B and C are more similar to each other than to OAS-TL A at the amino acid level. Hence, the strong cross-reactivity of the polyclonal antiserum, strongly detecting OAS-TL B and more weakly detecting the also abundant OAS-TL A protein (see below). Together, analyses of the genomic organization of T-DNA insertions, mRNA expression, and protein level corresponded to each other and demonstrated that the oastlA, oastlB, oastlC, and oastlAB lines were true null mutants of these genes.
Growth Phenotype of oastl Insertion Lines
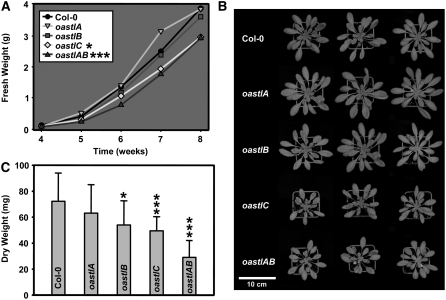
Arabidopsis Columbia (Col-0) and mutant lines oastlA, oastlB, oastlC, and oastlAB were grown in a growth chamber under optimized growth conditions. After 6 weeks, oastlA and oastlB were indistinguishable from the wild type. Lines oastlC and oastlAB were about the same size relative to each other but were significantly smaller compared with wild-type plants (Figures 2A and 2B). After 8 weeks, they reached only 75% of fresh weight compared with the wild type. This retardation of growth was observed during the entire growth period. Lines oastlC and oastlAB flowered later than the wild type, and oastlAB never reached wild-type size (data not shown). When grown under hydroponic conditions, the aerial parts showed the same phenotype, while no significant differences were noticed for the root systems. Root growth was also monitored for 2 weeks on agar plates, and no differences could be detected for either root length or branching pattern (data not shown). With respect to dry weight, rosettes of the oastlA mutant were not different from those of the wild type (Figure 2C). A trend to less weight was noted for oastlB, but measurements from 20 plants were only of weak statistical significance. Lines oastlC and even more so oastlAB clearly had less dry weight and confirmed the visible phenotype.
Figure 2.
Growth Phenotype of oastl Insertion Lines.
All plants were grown on soil in a growth chamber under short-day conditions.
(A) Fresh weight of aerial parts. Mean values from five plants are shown. Growth retardation of oastlC and oastlAB was statistically significant from week 4 on.
(B) Top view of the wild type and oastl insertion lines.
(C) Dry weights of aerial parts (n = 20). Mean values ± sd are shown.
Asterisks indicate statistical significance (* P < 0.05 > 0.01; *** P < 0.001).
OAS-TL Activities Differ in Subcellular Compartments
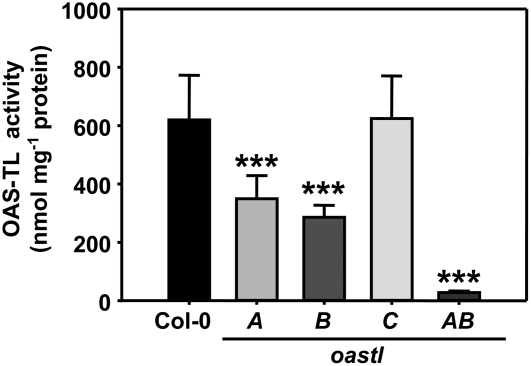
The relative contributions of OAS-TL activities were determined using the four loss-of-function insertion lines. Starting with Arabidopsis wild-type Col-0, OAS-TL activity measurements revealed a specific OAS-TL activity of 620 ± 150 nmol·min−1·mg−1 in protein extracts from leaves of 6-week-old plants (Figure 3). In plants lacking OAS-TL A or OAS-TL B protein, OAS-TL activity was on average reduced by 44 or 54%, respectively. The absence of OAS-TL C caused no measurable decrease in total OAS-TL activity. When both OAS-TL A and B were missing, OAS-TL activity was reduced by 22-fold. The residual activity of 5% in the oastlAB double mutant could be attributed to OAS-TL C, while contributions of the remaining OAS-TL isoforms were insignificant (see below).
Figure 3.
Total OAS-TL Enzyme Activity Varies from the Wild Type in oastl Insertion Lines.
Mean values ± sd refer to independent measurements from five plants. Total protein was extracted with extraction buffer A. *** P < 0.001.
The quantitative contribution of OAS-TL proteins in the three compartments and in the mutant lines was evaluated using a highly specific affinity purification scheme. SAT and OAS-TL are known to spontaneously form the CSC with a dissociation constant of 25 nM (Berkowitz et al., 2002). Moreover, SATs and OAS-TLs from Arabidopsis organelles and cytosol appear to form heterologous CSCs with comparable efficiency (Bogdanova and Hell, 1997; Jost et al., 2000). Therefore, cytosolic SAT5 from Arabidopsis was expressed in bacteria as a His tag fusion protein and immobilized on a nickel affinity column (see Supplemental Figure 2 online). Leaf protein extracts from 9-week-old wild-type plants were applied to the column, which retained 90 to 95% of total OAS-TL activity (Table 1). This suggests that nearly all OAS-TL proteins, independent of their subcellular localization, must have been recognized and retained by SAT5. OAS was used to specifically elute OAS-TL proteins from the column and yielded 38% of the total OAS-TL activity (Table 1). The OAS-TL purification was highly specific for OAS-TL proteins, and the pure fraction was apparently free of contamination (see Supplemental Figure 2 online).
Table 1.
Purification of OAS-TL Proteins from Wild-Type Leaf Extracts in Extraction Buffer B Using SAT Affinity Chromatography
| Fraction | Total Protein (mg) | Total Activity (μmol/min) | Specific Activity (μmol·min−1·mg−1) | Yield (%) | Purification Level |
|---|---|---|---|---|---|
| Crude extract | 257 | 488 ± 77 | 1.9 ± 0.3 | 100 | 1 |
| Flowthrough | 245 | 44 ± 3 | 0.18 ± 0.01 | 9 | 0.09 |
| Elution | 0.58 | 183 ± 19 | 315 ± 32 | 38 | 166 |
The purified OAS-TL proteins had a specific activity of 315 ± 32 μmol·min−1·mg−1 in the wild-type background (i.e., the OAS-TL protein fraction had been enriched by 166-fold) (Table 1). This value was comparable to the specific activities of purified recombinant OAS-TL A, B, and C from Arabidopsis (Wirtz et al., 2004). In summary, the remaining total activities found in the oastl mutants corresponded well with the amounts of the individual OAS-TL–like protein isoforms purified from wild-type plants by SAT affinity purification (Figure 3; see Supplemental Figure 2 online).
Abundance of Individual Isoforms and Posttranslational Modification of OAS-TL Proteins
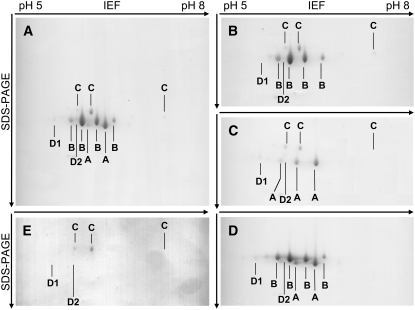
The purified OAS-TL fractions allowed us to precisely dissect whether cells of oastl mutants compensated for the loss of individual isoforms by increasing protein levels of one or more other OAS-TL isoforms in the same or other compartments. To determine the abundance of individual isoforms, purified OAS-TL protein fractions from 9-week-old Arabidopsis wild-type plants were separated by two-dimensional PAGE, using isoelectric focusing in the first dimension and SDS-PAGE in the second dimension. Surprisingly, more signals than expected were observed from the predicted number of OAS-TL–like proteins (Figure 4A). Identification of the signals by matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) analysis revealed that all belonged to true OAS-TL proteins. However, three different species for OAS-TL A, four different species for OAS-TL B, and three different species for OAS-TL C were detected. The OAS-TL A signal with the lowest isoelectric point was usually covered by an OAS-TL B spot on gels of wild-type protein and only became fully identifiable on gels of the oastlB mutant (Figure 4C). In addition, CysD1 and CysD2, which so far had been expressed as recombinant proteins from cDNA (Hatzfeld et al., 2000; Yamaguchi et al., 2000), were discovered as native proteins from Arabidopsis extract. Both proteins were present as single spots only. Their staining intensities were close to the detection limit and mirrored the comparatively low expression levels seen in microarray analyses (Zimmermann et al., 2004). The presence of different protein species of OAS-TL A, B, and C strongly suggested posttranslational modifications. However, electrospray ionization mass spectrometry analysis of their molecular masses, predicted sizes from OAS-TL cDNAs, and comparison with known modifications in plants allowed no conclusions with respect to their molecular identity at this point.
Figure 4.
Proteomic Profile of the OAS-TL Protein Family in Leaves of 9-Week-Old Arabidopsis Plants.
(A) Wild type.
(B) oastlA.
(C) oastlB.
(D) oastlC.
(E) oastlAB.
Native OAS-TL proteins were affinity-purified from leaf extracts using a SAT affinity column and identified by MALDI-TOF-MS analysis. CysD1 and CysD2 were not detectable in all Coomassie blue–stained gels, since their abundance was generally at the detection limit. IEF, isoelectric focusing.
When the intensities of the different species for each isoform were summed, the two-dimensional gel illustrated that OAS-TL A (23%) and OAS-TL B (65%) constituted the most abundant OAS-TLs, OAS-TL C (11%) had considerably lower levels, and CysD1 (1%) and CysD2 (0.25%) played even smaller roles in terms of protein abundance. Taking into account the specific activities of OAS-TL A, B, and C (Wirtz et al., 2004), these ratios corroborated the activity decreases observed in the oastlA, oastlB, and oastlC mutant lines (Figure 3). This, together with the specific activity of purified OAS-TLs (Table 1) that was comparable to the activities of recombinant enzymes (Wirtz et al., 2004), also suggested that the observed posttranslational modifications apparently had little effect on the measurable enzymatic activities in vitro. A possible function could be stabilization to enhance the half-life of the proteins.
Next, OAS-TL fractions from leaves of the single mutants and the oastlAB double mutant were analyzed by two-dimensional PAGE (Figures 4B to 4D). To allow comparison of staining intensities between gels of the four mutants, the Coomassie blue–stained gels were loaded with similar elution volumes from the purification column. This resulted in very weak signals of the OAS-TL C species, particularly those with the highest isoelectric point, as well as CysD1 and CysD2. However, the presence of these proteins was independently verified on other gels. No signals of OAS-TL A, B, and C could be detected in the corresponding single and double mutants independent of their posttranslational modifications. Moreover, no signal of any OAS-TL isoform or their modified protein species changed its intensity in comparison with the wild type when OAS-TL A, B, or C was absent. The same was true for the oastlAB mutant. The abundance of the five OAS-TL proteins that bound to SAT correlated well with the relative amounts of mRNAs in leaves and roots at this developmental stage in the literature (Hell et al., 1994; Hesse et al., 1999; Hatzfeld et al., 2000; Yamaguchi et al., 2000) and in a public microarray database (Zimmermann et al., 2004).
Properties of the OAS-TL–Like Proteins CysC1, CS26, CysD2, and CS-Like with Respect to Cys Synthesis
The remaining OAS-TL–like proteins were of minor abundance or not detected at all. Nevertheless, if they catalyzed true OAS-TL reactions, they could possibly provide residual amounts of Cys in the affected compartments. To ensure that no possible compensatory changes in the not-detected isoforms had been overlooked, first the expression of the genes encoding CysC1, CS26, and CS-like protein was determined. In the four oastl mutants, little or no differences were detected in the mRNA levels of any of these genes in comparison with Col-0 wild type (see Supplemental Figure 3 online).
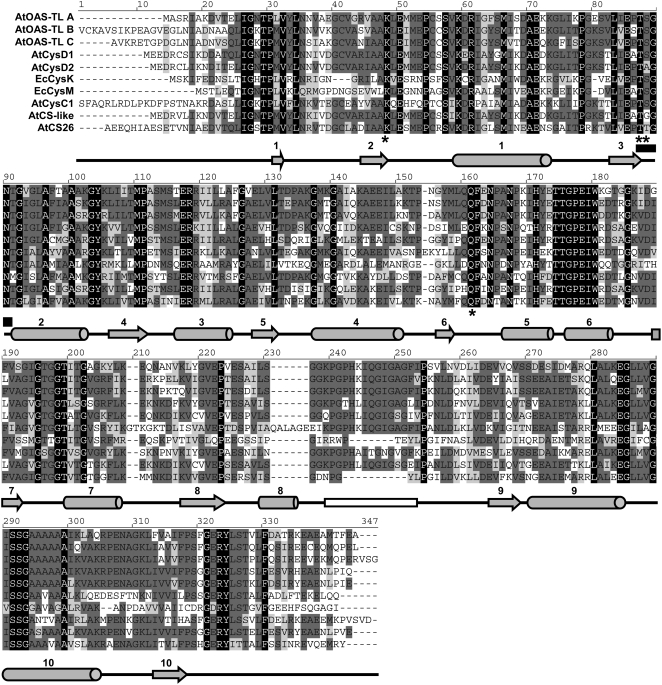
In addition, the CysC1, CS26, and CS-like proteins were evaluated with respect to enzymatic properties and interaction with SAT, a feature that has been suggested as a defining condition for enzymatically true OAS-TL proteins (Droux, 2004). Mitochondrial CysC1 had been shown to be a CAS and assumed to not catalyze an OAS-TL reaction in vivo (Hatzfeld et al., 2000; Yamaguchi et al., 2000). This protein was thus not relevant for the assessment of Cys synthesis and was not expected to interact with SAT. Next, amino acid sequences of all Arabidopsis members of the family of OAS-TL–like proteins were aligned with the catalytically and structurally well-investigated Escherichia coli OAS-TL proteins CysK and CysM (Figure 5) (Rabeh and Cook, 2004). The comparison revealed that CS26, predicted to reside in the plastid, did not have the β8a-β9a loop, a domain necessary for interaction with SAT (Bonner et al., 2005). The putatively cytosolic CS-like protein and the mitochondrial CysC1 showed substantial amino acid changes within this loop, strongly suggesting that these three members of the OAS-TL–like family cannot be purified by interaction with SAT. In agreement with these findings, E. coli CysM, using thiosulfate instead of sulfide as substrate, lacked the β8a-β9a loop and showed no interaction with E. coli SAT (Zhao et al., 2006). Together, these data strongly suggested that cytosol as well as plastids and mitochondria in the T-DNA mutants were depleted of OAS-TL enzymatic activities. This conclusion further implied that sulfur compounds could be efficiently transported between cytosol and organelles.
Figure 5.
Catalytic and Interaction Domains of Arabidopsis and E. coli OAS-TL–Like Proteins.
Sequence alignment and secondary structure prediction of cytosolic (OAS-TL A, CysD1, CysD2, and CS-like), plastid (OAS-TL B and CS26), mitochondrial (OAS-TL C and CysC1), and E. coli (CysK and CysM) proteins were created with Vector NTI9. Identical residues in all sequences are shown in white on black background, while the declining degree of conservation is indicated by the degree of lighter gray background. Below the alignment, α-helices are shown by tubes and β-sheets by arrows. A black rectangle marks the Asn loop (part of the catalytic center), and a white rectangle marks the β8a-β9a loop (SAT interaction). Important residues for substrate binding and formation of the catalytically active site are marked with asterisks.
Impact of Altered Cys Synthesis Capacity on Sulfur Metabolism–Related Compounds
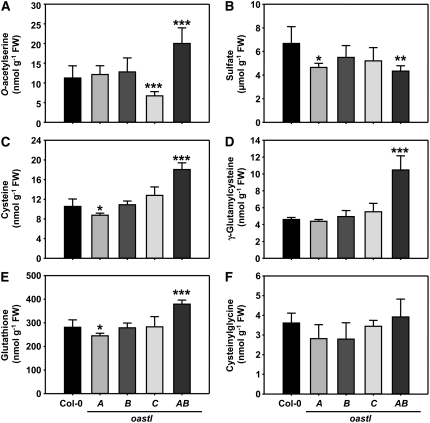
In wild-type plants, the steady state concentrations of metabolites related to primary sulfur metabolism are in the order sulfate ≫ GSH ≫ Cys, OAS > γ-glutamylcysteine (γEC), cysteinylglycine (≫ indicating >10-fold differences on a fresh weight basis). Within this general range, a number of significant changes of steady state concentrations were observed in the four oastl insertion lines (Figure 6). First, and rather unexpectedly, neither Cys nor GSH levels were much decreased in either of the single mutants, although OAS-TL A and B were shown to each contribute about half of total cellular Cys biosynthetic capacity. The concentrations of both thiols were not altered at all in oastlB and decreased only slightly in oastlA at low statistical significance. Second, the OAS level in oastlC was only about half of the concentration in the wild type and oastlA and oastlB. Third, the oastlAB double mutant had increased contents of Cys (almost twofold), γEC (about twofold), GSH (+30%), and OAS (almost twofold) compared with the wild type. Only the assumed degradation product of GSH, cysteinylglycine, and sulfate contents remained unaffected. In the oastlAB mutant, the mitochondrial OAS-TL C protein together with the minor contribution of CysD1 presumably constituted the only remaining capacity for Cys synthesis in the cell. The changes in OAS concentrations were in agreement with mitochondria being the major site of SAT activity (see below) (Ruffet et al., 1995; Droux, 2003) and with the hypothesized dependence of SAT activity on interaction with OAS-TL (see below) (Droux et al., 1998; Wirtz and Hell, 2007). The increase in Cys-related compounds may thus reflect an overcompensation of mitochondrial Cys synthesis by the CSC and possibly a lack of metabolite exchange or communication with the cytosol and plastids. It should be noted in this context that GSH synthesis is split between the latter compartments in Arabidopsis (Wachter et al., 2005). Finally, in contrast with the changes described before, the fluctuations in sulfate contents in oastlA and oastlAB mutants were at the limit of statistical significance and, considering the high levels of sulfate compared with the other compounds, probably physiologically insignificant. Nitrate contents in the four oastl mutants were also unchanged, as were the total contents of carbon, nitrogen, and sulfur (see Supplemental Figure 4 online).
Figure 6.
Sulfur-Related Metabolites in oastl Insertion Lines.
Metabolites were extracted from leaves of the wild type (Col-0) and indicated oastl mutants and quantified by HPLC. Mean values ± sd are shown for independent analyses from five plants of each genotype. Asterisks indicate statistically significant differences compared with wild-type plants (* P < 0.05 > 0.01; ** P < 0.01 > 0.001; *** P < 0.001). FW, fresh weight.
Catalytic Reactions Upstream of Cys Synthesis in oastl Insertion Lines
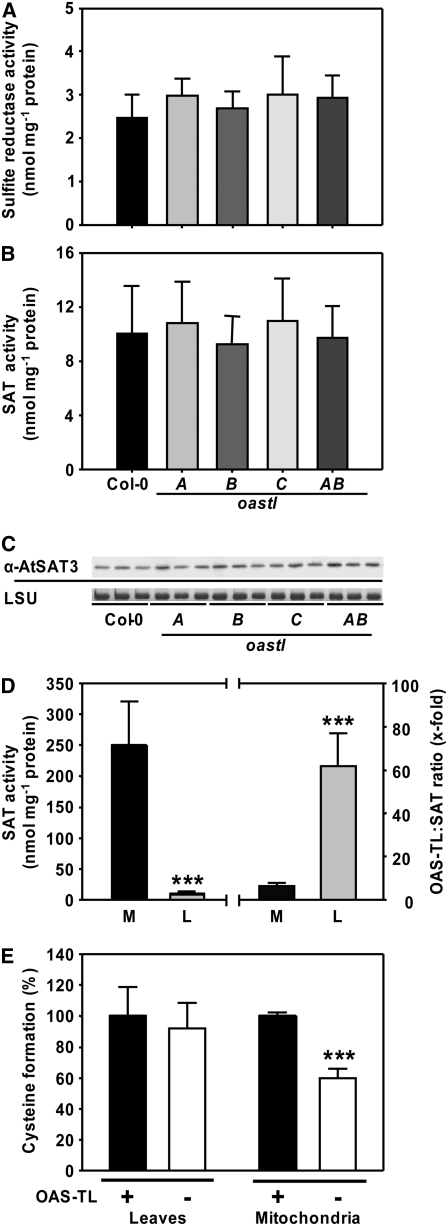
The synthesis of Cys represents the convergence of carbon/nitrogen metabolism via Ser with sulfur metabolism and is fueled by sulfide generation through assimilatory sulfate reduction. The activities of important reactions of both branches of the pathway immediately upstream of OAS-TL catalysis were assayed in vitro in the four oastl mutants (Figures 7A and 7B). The total activities of neither SAT nor sulfite reductase were changed. Mean values for specific activities of sulfite reductase were slightly increased in the mutants but not statistically significant (Figure 7A). Since SAT, like OAS-TL, resides in plastids, mitochondria, and cytosol, the assumed major isoform, mitochondrial SAT3, was evaluated using a specific polyclonal antiserum (Figure 7C). However, SAT3 protein levels were not affected in the oastl mutants, suggesting that the changed fluxes of sulfide and Cys in these plants did not affect the levels of both enzymes that provide the substrates for the OAS-TL reaction. In organellar oastl insertion lines, the absence of OAS-TL resulted in the loss of CSC formation in the respective compartment. It has to be noted at this point that an inactivation of organellar SAT in vivo by the lack of CSC formation would not be observable in the in vitro SAT assay, due to the reactivation of the respective SAT by the high excess of OAS-TLs (Ruffet et al., 1995; Droux, 2003) from other compartments during extraction.
Figure 7.
Specific Activities of SAT and Sulfite Reductase.
(A) and (B) Mean values ± sd from five independent protein extractions of the wild type and oastl insertion lines are shown.
(C) An immunoblot loaded with protein from three extractions of leaves of each genotype was detected with an At SAT3 polyclonal antiserum. Staining of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase protein (LSU) with Coomassie blue in the same samples confirmed equal loading in the individual lanes.
(D) Specific SAT activity in crude extracts of leaves (L) and isolated mitochondria of cell cultures (M). Mean values ± sd from three independent protein extractions are shown (left). The ratio of OAS-TL to SAT in the respective fractions is shown at right. *** P < 0.001.
(E) Conversion of OAS, synthesized by SAT in the assay, to Cys in the absence (−) and presence (+) of a 100-fold excess of OAS-TL activity achieved by the addition of recombinant OAS-TL A. The capacity for Cys formation in the presence of recombinant OAS-TL activity was set to 100% in crude extracts of leaves (L) and isolated mitochondria (M). In contrast with endogenous total OAS-TL, the mitochondrial OAS-TL failed to convert all SAT-synthesized OAS to Cys, which confirms the low mitochondrial OAS-TL:SAT activity ratio (see [D]).
The significance of mitochondrial Cys synthesis implied by oastlC and oastlAB mutants and the role of OAS levels as an indicator for the activity of sulfate assimilation (Hirai et al., 2003; Saito, 2004) prompted us to probe SAT activities in mitochondria of Arabidopsis. The specific SAT activity of mitochondrial protein extracts was 25-fold higher than in the crude plant extracts (Figure 7D). The higher specific SAT activity in mitochondria of Arabidopsis indicated a similar localization of SAT activities to that observed in pea (Ruffet et al., 1995), in which almost 80% of total SAT activity was found in the mitochondria. The OAS-TL activity excess was ∼60-fold in crude extracts of plants, while the OAS-TL:SAT ratio was only 7:1 in the purified mitochondria of Arabidopsis cell cultures (Figure 7D). The low OAS-TL:SAT activity ratio indicated an insufficient capacity for the conversion of OAS to Cys in mitochondria, since in vitro a 300-fold excess of OAS-TL:SAT activity is necessary to ensure the full conversion of OAS to Cys (Droux et al., 1998). Indeed, when mitochondrial protein extracts were assayed for SAT activity in the absence or presence of a >100-fold excess of OAS-TL activity, the formation of Cys from OAS was almost doubled, confirming that also in Arabidopsis mitochondria, at least from cell culture, the CSC could not reach maximal Cys production under native enzyme ratios (Figure 7E). The addition of OAS-TL to crude extracts of plants caused no such increase, due to an already high excess of OAS-TL activity in cytosol and plastids. The low OAS-TL:SAT activity ratio furthermore suggested that under wild-type conditions, the mitochondria are the major site for the production of OAS rather than Cys. Most likely, part of the OAS generated in mitochondria will support Cys formation in cytosol or plastids (see Figure 9 below).
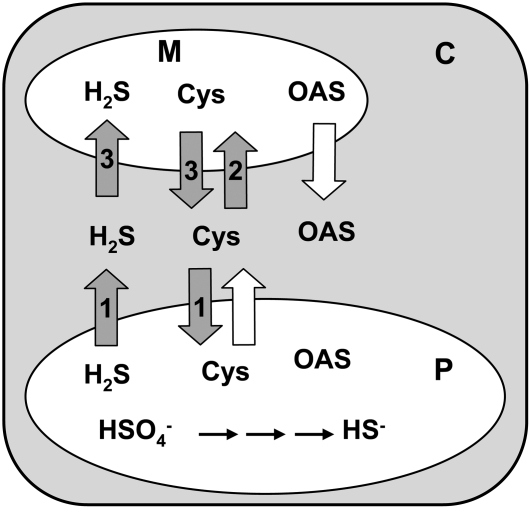
Figure 9.
Transport Processes between Cytosol, Plastid, and Mitochondrion in the oastl Insertion Lines.
Transport of sulfide and Cys (dark arrows containing numbers) is indicated by the survival of the following mutant lines: 1, oastlB; 2, oastlC; 3, oastlAB. The white arrows show the predicted transport of OAS and Cys based on results obtained for oastlA, oastlC, and oastlAB and the low OAS-TL:SAT activity ratio in mitochondria of wild-type Arabidopsis. C, cytosol; P, plastid; M, mitochondrion.
Incorporation of Radioactively Labeled Sulfate into Cys
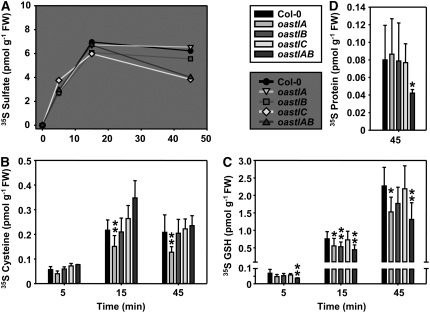
The above findings strongly suggested differential roles of Cys synthesis in the cytosol and organelles. Feeding experiments were performed in order to reveal the underlying fluxes of sulfur from assimilatory sulfate reduction into Cys in vivo. Leaf pieces from Arabidopsis wild-type and mutant plants were floated on half-strength Hoagland medium containing radioactively labeled 35SO42− to monitor the incorporation of labeled sulfate and flux into Cys, GSH, and protein. Incorporation was measured after 5 and 15 min of incubation with 35SO42−. After another 30 min of incubation on medium without 35SO42−, another sample was taken as a 45 min value. Comparable and linear uptake of 35SO42− into the mutants and wild-type leaves was observed between 0 and 15 min, followed by a slight decrease during the 30 min chase due to the consumption of sulfate by assimilatory reduction (Figure 8A). After 45 min, the highest 35S label was found in sulfate (4 to 6 pmol/g fresh weight), followed by GSH (1.5 to 2.5 pmol/g fresh weight), Cys (0.15 to 0.25 pmol/g fresh weight), and protein (0.04 to 0.08 pmol/g fresh weight). Apart from sulfate, the bulk of the label under these conditions went into GSH and not into protein. The steady state of 35S label was not reached in GSH after 45 min. By contrast, Cys appeared to have a much higher turnover, because the label showed no further increase after 15 min (Figures 8B and 8C). Under these conditions, feeding did not interfere with primary sulfur metabolism, since only traces of the cellular Cys and GSH pools were labeled (cf. Figures 6 and 8). [35S]Cys of 0.22 ± 0.04 pmol/g fresh weight after 15 min corresponded to 1/50,000 of the total Cys pool, and [35S]GSH amounted to 0.76 ± 0.20 pmol/g fresh weight, representing 1/370,000 of the total GSH pool after 15 min.
Figure 8.
Incorporation of 35SO42− into Cellular Sulfur Compounds in Vivo.
Leaf pieces of the wild type and the four indicated oastl mutant genotypes were incubated with 35SO42−-spiked half-strength Hoagland solution for 15 min and subsequently kept on the same medium without radiolabel until 45 min after the start of the experiment. Samples were taken at the indicated time points. Sulfate, thiols, and proteins were separated by HPLC or specific precipitation, and incorporated 35S was quantified by scintillation counting. FW, fresh weight. Mean values ± sd are shown. * P < 0.05 > 0.01; ** P < 0.01 > 0.001.
The oastlA mutant was the only one of the four mutants with a significantly reduced incorporation of radioactively labeled sulfur into Cys. Label in GSH was also reduced after 15 and 45 min. This observation fits with the slightly reduced steady state concentration of Cys (Figure 6) and suggested that OAS-TL A is more important for the bulk of Cys formation than the other isoforms. However, this reduction of Cys biosynthesis in the oastlA mutant was not limiting for plant growth. Remarkably, the chloroplast localization of OAS-TL B would have suggested that the oastlB mutant was actually most affected with respect to the short-term incorporation of 35S into Cys, but in agreement with the unchanged steady state concentrations of thiols this was not the case, except for GSH at time point 15 min. Most notably, the results imply that sulfide must have been able to rapidly cross the plastid envelope into the cytosol and probably also into mitochondria.
It seemed reasonable to assume that the highly abundant OAS-TL A and OAS-TL B proteins in cytosol and plastid mutually compensated for their absence in the oastlA and oastlB mutants, respectively. This option was eliminated in the oastlAB mutant, which was retarded in growth and relied almost entirely on Cys synthesized by OAS-TL C in mitochondria. Remarkably, like the steady state concentrations, the flux into Cys was not impaired in this mutant at all. Therefore, lack of Cys per se was unlikely to be responsible for the reduced incorporation of 35S into GSH and protein (Figure 8D), although it cannot be excluded that the latter is indirectly connected to the reduced growth rate. The reduced incorporation of 35S into GSH and protein may be attributed to an insufficient transport of Cys across the mitochondrial envelope into the cytosol and the plastids, at least during the short labeling pulse of the experiment. In contrast with oastlAB, the oastlC mutant still contained OAS-TL A and B and showed no effect on the incorporation of label into thiols and protein. The observed reduced OAS concentrations in oastlC were apparently not limiting for incorporation of label into Cys during the experiment. Therefore, the retarded growth of oastlC cannot be linked to a limited capacity for Cys synthesis, GSH, or protein. The phenotype supposedly had a different cause than the phenotype of oastlAB, such as decreased SAT activation due to the lack of OAS-TL for CSC formation or a limitation of Cys import into the mitochondria of the oastlC line. In summary, the flux experiments confirmed the existence of efficient transport processes for sulfide and Cys across the organellar membranes and the ability to compensate for the loss of Cys synthesis in one of the three compartments. However, Cys synthesis in mitochondria and also the cytosol was clearly more important than the site in plastids.
DISCUSSION
Plastids, and in particular chloroplasts, are generally viewed as the major site of Cys synthesis and export Cys or possibly other thiols to the cytosol and eventually mitochondria (Hell, 1997; Leustek and Saito, 1999; Kopriva, 2006). Directed transport of Cys across the organelle envelopes was simply assumed and thought to be performed by transport proteins with limited specificity, such as the transporters involved in the uptake of amino acid at the plasmalemma (Miranda et al., 2001; Fischer et al., 2002). However, direct evidence for the transport of Cys between cytosol and compartments is scarce. Such an exchange was proposed from an analysis of transgenic potato (Solanum tuberosum) with decreased plastid OAS-TL activity (Riemenschneider et al., 2005) but was found less likely in transgenic tobacco expressing an inactivated SAT protein that was able to form a heterologous CSC in the cytosol (Wirtz and Hell, 2007). In yeast, the Bap3 protein was reported in a proteomics approach to reside in the inner mitochondrial membrane (Reinders et al., 2006) and has been found to be involved in cellular Cys uptake in genetic experiments (Düring-Olsen et al., 1999). Even less is known about the transport of GSH between compartments of the plant cell. Only one report is available that describes the uptake of [35S]GSH into isolated chloroplasts of wheat (Triticum aestivum) (Noctor et al., 2002).
New insight into the open questions of the compartmentation of Cys synthesis in plant cells was provided here using affinity purification of the OAS-TL–like protein family of Arabidopsis in combination with T-DNA insertion mutants of the OastlA, OastlB, and OastlC genes encoding cytosolic, plastid, and mitochondrial OAS-TLs. The three T-DNA insertion lines and a double mutant of oastlA and oastlB were shown to be null mutants with respect to mRNA and the encoded proteins. Affinity purification and two-dimensional PAGE demonstrated that no compensatory changes in the OAS-TL protein profile occurred in the mutants. The profiling of five of the eight known OAS-TL–like proteins using SAT affinity purification confirmed structural studies that predicted the consensus β8a-β9a loop in OAS-TL–like proteins as necessary for binding to SAT (Bonner et al., 2005; Francois et al., 2006). Amino acid changes in this loop resulted in an apparent inability of the OAS-TL–like proteins CysC1 (=CAS), CS26, and CS-like to stably interact with SAT and correlated with a lack of OAS-TL enzymatic activity (Hatzfeld et al., 2000; Yamaguchi et al., 2000; Bonner et al., 2005; Francois et al., 2006). An OAS-TL–like protein from spinach mitochondria was found to be a CAS with an OAS-TL side activity and led to the conclusion that this species harbors no true OAS-TL protein in mitochondria (Warrilow and Hawkesford, 2000). Thus, CAS might be able to replace OAS-TL in mitochondria, at least in spinach. While the presence of other OAS-TL–like proteins in spinach mitochondria cannot be ruled out (Yamaguchi et al.,1998), enzymatically defined mitochondrial OAS-TLs have been demonstrated in D. innoxia and Arabidopsis, suggesting that CAS probably has no function for Cys synthesis in these species (Kuske et al., 1994, 1996; Hesse et al., 1999; Jost et al., 2000).
The OAS-TL A–deficient mutants oastlA and oastlAB still carried CysD1 and CysD2. Therefore, these mutants could potentially carry out some Cys biosynthesis in the cytosol. But CysD1 was not increased in its already very low abundance, restricting any residual OAS-TL activity in the cytosol of the oastlA and oastlAB mutants to the very minor amount of this isoform. The weak potential CysD1 contribution in vivo is also supported by its low specific OAS-TL activity and its 10 times higher Km values for OAS and sulfide compared with true OAS-TLs, although it should be cautioned that the enzyme was assayed in fusion with glutathione S-transferase (Yamaguchi et al., 2000; Wirtz et al., 2004). Any substantial catalytic OAS-TL activity of cytosolic CysD2, but also CS26 and the CS-like protein, most likely can be excluded due to an exchange of Ser at position 88 in the Asn loop that is part of the OAS-TL catalytic center (Rabeh and Cook, 2004). In fact, Bonner et al. (2005) demonstrated that mutation of Ser-88 in At OAS-TL A resulted in a loss of catalytic OAS-TL activity; accordingly, a rather low specific OAS-TL activity of recombinant CysD2 was reported (Yamaguchi et al., 2000). Therefore, the OAS-TL–like proteins CysC1, CysD2, CS26, and CS-like could be eliminated as contributors to Cys synthesis in their respective cell compartments. For the mutants missing OAS-TL B or OAS-TL C, it could be concluded that the plastids or the mitochondria, respectively, were completely deprived of Cys synthesis, while in the mutants missing OAS-TL A only insignificant residual activity was left in the cytosol.
This constellation of Cys synthesis activities in the oastl insertion lines revealed several implications that help elucidate the relations of the locations of Cys synthesis in the Arabidopsis cell. For one, the role of plastid Cys synthesis is less important than assumed, despite taking place at the site of assimilatory sulfate reduction and sulfide formation. Cys synthesis in plastids was not a prerequisite for the fixation of reduced sulfur (Hell, 1997; Leustek et al., 2000; Saito, 2004), nor did it affect the rate of sulfate reduction according to pulse-chase [35S]sulfate labeling experiments. Lack of plastid Cys synthesis could be completely compensated for by the other compartments with respect to growth phenotype, steady state thiol concentrations, and flux of reduced sulfur into Cys and protein. Next, the significance of cytosolic OAS-TL A has been underestimated. It constituted 23% of total OAS-TL protein and 44% of total OAS-TL activity in leaf and ∼80% in roots (data not shown). Loss of OAS-TL A resulted in the only observable decrease of flux into Cys in leaf feeding. Nevertheless, the oastlA mutant line had no growth phenotype. Third, mitochondrial OAS-TL is much more important than assumed, at least in Arabidopsis. In an oastlAB mutant, OAS-TL C was sufficient for sustained growth, although at the cost of growth rate. In turn, the loss of OAS-TL C caused growth retardation.
This mutant phenotype strongly suggested mitochondrial Cys synthesis as a major control point. Since thiol concentrations and incorporation rates into Cys and protein were unchanged, the growth retardation of oastlC was not a consequence of limited Cys production. One explanation could be an insufficient import of Cys across the mitochondrial envelope. While this possibility cannot be ruled out, it seems unlikely, because large amounts of Cys must be able to pass from the mitochondria to the cytosol in the oastlAB mutant line. More reasonably, the phenotype of oastlC resulted from the deregulation of SAT activity in mitochondria. Mitochondria are the major site of OAS synthesis but have an extremely low ratio of OAS-TL to SAT activity of ∼4:1 compared with cytosol (200:1) and chloroplasts (300:1) in pea, the only plant that had been analyzed for both enzymes so far (Ruffet et al., 1995; Droux, 2003). In this study, the ratios were confirmed for Arabidopsis mitochondria and whole protein extract. SAT activity was described to depend at least in part on a high excess of OAS-TL protein as part of the regulatory circuit of the CSC sensing system (Droux et al., 1998; Berkowitz et al., 2002; Sirko et al., 2004; Wirtz and Hell, 2007), and this could indeed be shown for Arabidopsis mitochondria as well. Thus, any change in the abundance of mitochondrial OAS-TL has strong effects on OAS formation, possibly in the entire cell, because ∼80% of SAT activity was in mitochondria (Ruffet et al., 1995; Droux, 2003). Correspondingly, the steady state concentration of OAS was clearly lowered in oastlC. The elevated level of OAS in the oastlAB mutant, in which mitochondrial OAS-TL C contributed basically all of the Cys in the cell, might be interpreted as an enhanced activation of mitochondrial SAT. In this mutant, more CSC-stabilizing sulfide entered the mitochondria (because plastid and cytosolic Cys synthesis were lacking) and futile export of OAS into the cytosol and plastids took place. The well-documented feedback inhibition of several SAT proteins by Cys was probably less important for flux regulation. An elegant hypothesis suggested that in Arabidopsis, cytosolic SAT is highly feedback-sensitive to Cys, whereas plastid and mitochondrial SATs are much less susceptible (Noji et al., 1998). Indeed, mitochondrial SAT in Arabidopsis showed 50% inhibition of maximal specific activity at Cys concentrations between 50 and 100 μM, while cytosolic SAT reached 50% inhibition at 2 to 10 μM Cys (Noji et al., 1998). Since Cys contents in the oastl mutants showed only 1.5-fold increases, at most the changes in feedback inhibition of mitochondrial SAT appeared to be less important.
A remarkable result of the analysis of the oastl insertion lines was the nearly constant overall concentrations of Cys and GSH. While compartment-specific differences in the steady state levels of thiols cannot be excluded entirely, it appears to be important for the cell to maintain these levels, even when the incorporation rate is reduced, such as in the oastlA mutant. Reduction of cytosolic and plastidial OAS-TL activities in potato by a targeted RNA interference approach does also not result in decreased total Cys and GSH levels (Riemenschneider et al., 2005). It seems reasonable to assume that the unaffected compartments provided Cys for sites where OAS-TL proteins had been eliminated. These compensations in the oastl mutants took place without noticeable changes in the pattern of OAS-TL and SAT proteins. The effector-driven regulation of the CSC provides an elegant explanation for this observation. Loss of OAS-TL B activity, for instance, meant that more sulfide left the plastid into the cytosol, given that the rate of sulfate reduction was the same in the oastlB mutant. The latter was apparently the case according to 35S incorporation rates. Increased availability of sulfide in the cytosol and likely also the mitochondria of this mutant stabilized the CSC in these compartments (Wirtz and Hell, 2007), resulting in enhanced SAT activity and OAS formation. Similar scenarios apply to the other mutant lines, supporting once again the CSC regulatory model (Hell and Hillebrand, 2001; Wirtz and Hell, 2006).
The absence of Cys synthesis in the plastid, mitochondrion, and most likely also the cytosol held important implications for the transport of Cys, sulfide, and OAS (Figure 9). In contrast with the suggestion by Lunn et al. (1990) and with the lack of any direct evidence, the viability of mutants with complete absence of Cys synthesis in the compartments with their own protein biosynthesis can only be explained by efficient transport processes between the cytosol and both organelles. All four mutants were fully viable, at least under growth chamber conditions, with oastlC and oastlAB showing significant, but not dramatic, growth retardation. The lack of OAS-TL B in plastids of the oastlB mutant, therefore, meant that sulfide, generated in the plastids by assimilatory sulfate reduction, must have been available in sufficient amounts for compensatory Cys synthesis in the cytosol and mitochondria (shown by the oastlAB mutant). These data demonstrate that Cys had been imported into the plastid of oastlB lines for protein biosynthesis, synthesis of GSH, and other basic processes. In addition, Cys also must have been able to cross the envelope of mitochondria in both directions (1) to provide Cys for mitochondrial processes in oastlC mutants and (2) to supply the cytosol in the oastlAB mutant. Cys most likely left the plastids in the oastlA mutant, although an oastlAC double mutant would be required to rule out the possibility that mitochondrial OAS-TL C compensated completely for Cys in the cytosol without any contribution from plastid Cys synthesis. Similar transport processes were also assumed to take place in potato, since a transgenic potato line with reduced plastid OAS-TL activity exhibited higher thiol levels than the wild type (Riemenschneider et al., 2005).
The RNA interference approach used for potato may support the results from the Arabidopsis T-DNA lines but probably was less specific with respect to OAS-TL isoforms and differed in the extent of elimination of OAS-TL activities in the targeted compartments. An explanation for the accumulation of OAS in the oastlAB mutant may be that OAS left the mitochondria, its primary site of production, to end up in the cytosol, where it could not be converted into Cys due to the lack of OAS-TL A. In accordance, the ratio of OAS-TL to SAT activity in wild-type mitochondria of Arabidopsis was low, which suggested that not all OAS produced in mitochondria could be converted to Cys in the same compartment (Figure 7). In this respect, the potential toxicity of sulfide for the mitochondrial respiratory chain has to be considered. The rather mild but significant growth retardation of the oastlC and oastlAB mutants could be a result of lowered sulfide detoxification due to the lack of OAS-TL C or elevated sulfide concentrations in mitochondria due to the lack of OAS-TL A and OAS-TL B, respectively. Indeed, concentrations as low as 0.002 mM sulfide may negatively affect root respiration and nutrient uptake. In rice (Oryza sativa) roots, sulfide-induced inhibition of respiration could directly be explained by a reaction of sulfide with the heme group of cytrochrome c oxidase (Allam and Hollis, 1972). By contrast, shoot respiration was not susceptible to exposure to elevated atmospheric H2S levels (Maas et al., 1988). Total carbon, nitrogen, and sulfur analyses (see Supplemental Figure 4 online) showed comparable amounts of total N and S in all oastl insertion lines, suggesting no significant reduction of nutrient uptake by the root. The morphology and growth of roots were also not affected in oastlC and oastlAB plants when grown in hydroponic culture. Compared with other plant species, Arabidopsis and Brassica oleracea have 10-fold higher uptake rates for atmospheric H2S (∼0.3 nmol·g−1 fresh weight·s−1), indicating a high resistance toward sulfide (De Kok et al., 1997). Therefore, the slow growth phenotype of oastlC and oastlAB was most likely not a consequence of inhibited mitochondrial respiration in the root or the shoot.
Alternative to the proposed transport processes, reduced sulfur could be imported into plastids or mitochondria as GSH, Met, or protein-bound sulfur. The latter would provide too small transport rates, but efficient import of GSH into plastids is likely (Pasternak et al., 2008). Import would require degradation, but γ-glutamyltransferases for degradation of GSH or γEC at least in Arabidopsis are localized at the outer plasmalemma surface or in the vacuole (Storozhenko et al., 2002; Grzam et al., 2007; Martin et al., 2007; Ohkama-Ohtsu et al., 2007a, 2007b). Degradation of Met has been reported, but the enzyme is localized in the cytosol, produces Ile and S-methylcysteine, and is assumed to have very low activity in vivo (Rebeille et al., 2006). Moreover, consumption of these thiol compounds would require anabolism and catabolism in the same compartment. Thus, import of Cys seems by far the more likely mechanism.
Membrane transport of sulfide may occur as H2S, as suggested (Wirtz and Hell, 2007). At physiological pH values, a substantial part of the sulfide equilibrium is in the uncharged form, while HS− forms the actual substrate of the OAS-TL reaction (Wirtz et al., 2004). However, the rates of possible membrane diffusion of H2S are unknown and might require a currently unknown active membrane transport process in metabolic situations of high demand.
The lesson from analysis of the OAS-TL protein family in Arabidopsis is functional redundancy between the compartments of Cys synthesis, due to membrane transport and relatively equal contributions despite strong differences in protein abundance. Plastid Cys synthesis is dispensable, but the mitochondrial site is important for the entire metabolism of reduced sulfur in the cell.
METHODS
Plant Genotypes and Growth Conditions
All experiments were performed using Arabidopsis thaliana ecotype Col-0 as the wild-type control, since all mutants were derived from or backcrossed to this accession. Seeds for oastlB (N521183) and oastlC (N500860) plants were obtained from the SALK collection (Salk Institute Genomic Analysis Laboratory). The oastlA mutant, derived from the T. Jack collection (Campisi et al., 1999), was isolated out of a pool of 10 plants (N19847) from the screening for the atToc33 mutant (Gutensohn et al., 2004) and then backcrossed with Col-0. All seeds were verified for homozygosity of the corresponding T-DNA insertion. Seeds were stratified on soil for 3 d at 4°C in the dark and transferred for germination to climate chambers. After 2 weeks, seedlings were transferred to individual pots and grown for another 6 to 9 weeks under short-day conditions (8.5-h light period). The light intensity was set to 100 μE·m−2·s−1, while the RH was kept at 50%. The temperature during the day and night changed from 22 to 18°C. For hydroponic cultures, seeds were germinated in Eppendorf tubes and placed on small boxes (0.25 L) as described by Tocquin et al. (2003) containing half-strength Hoagland solution [2.5 mM Ca(NO3)2, 2.5 mM KNO3, 0.5 mM MgSO4, 0.5 mM KH2PO4, 40 μM Fe-EDTA, 25 μM H3BO3, 2.25 μM MnCl2, 1.9 μM ZnSO4, 0.15 μM CuSO4, and 0.05 μM (NH4)6Mo7O24, pH 5.8 to 6.0]. After 16 d, four individual seedlings of each genotype were transferred to large pots containing 5 L of the same medium and grown for an additional 19 d. Growth media were exchanged every 2 weeks. Hydroponically grown plants were kept under long days (14-h light period) and otherwise the same conditions as soil-grown plants.
Molecular Cloning and Genomic Characterization
Standard molecular biology technologies, such as growth of bacteria, plasmid isolation, and PCR, were performed as described by Sambrook et al. (1989) according to Good Laboratory Practice standards. Genomic DNA was extracted from plants according to Dellaporta (1982). For selection and the genomic characterization of homozygous plants, a PCR screen was set up. Specific primers were designed for each gene to bind upstream and downstream of the predicted T-DNA insertion as well as one primer to bind in the left border region of the corresponding T-DNA. The following primers were used: TJLB (5′-GAACATCGGTCTCAATGCAAAAGGGGAAC-3′) for the T-DNA of the oastlA mutant, and LB_SALK (5′-GACCGCTTGCTGCAACTCTCTCAGG-3′) for that of both oastlB and oastlC. The gene-specific primers were A_for (5′-CTCACAAGATTCAAGGGATAGGA-3′), A_rev (5′-GTCATGGCTTCCGCTTCTTTC-3′), B_for (5′-TGACTTCTCGCCACCGTCCT-3′), B_rev (5′-TGCAACACAGCCCTTGACTACA-3′), C_for (5′-CGATGATCATGGCTTCAAGG-3′), and C_rev (5′-CGATGAATGCTAGGCCAATACCCGTG-3′). Amplified products were sequenced to verify the insertion site.
Determination of Metabolites
Hydrophilic metabolites were extracted from leaves of Arabidopsis plants according to Wirtz and Hell (2003). Thiols and amino acids were quantified after derivatization with monobromobimane (Calbiochem, EMD Chemicals) and AccQ-Tag reagent (Waters), respectively. The derivatization procedure and separation of thiol derivatives were performed as described by Wirtz et al. (2004) using the same HPLC system. Anions were separated and quantified after 10-fold dilution in water according to Wirtz and Hell (2007).
Determination of Enzymatic Activities and Immunological Detection of Proteins
Aliquots of 200 mg of leaf material were powdered in liquid nitrogen for small-scale extraction of proteins. The proteins were extracted at 4°C for 15 min with frequent shaking in 0.5 mL of extraction buffer A (50 mM HEPES-KOH, pH 7.4, 10 mM KCl, 1 mM EDTA, 1 mM EGTA, 10% [v/v] glycerin, 10 mM DTT, and 0.5 mM phenylmethylsulfonyl fluoride [PMSF]). Cell debris was collected by centrifugation at 4°C for 10 min and 25,000 g, and the resulting supernatant was applied to a NAP-5 column (GE Healthcare) that was equilibrated in resuspension buffer (50 mM HEPES-KOH, pH 7.5, and 1 mM EDTA). Proteins were eluted with 1 mL of resuspension buffer, supplied again with 2 mM DTT and 0.5 mM PMSF, and quantified as described by Bradford (1976) using BSA as a standard. The enzymatic activity of OAS-TL was determined according to Gaitonde (1967) at 25°C in a total volume of 0.1 mL containing 100 mM HEPES, pH 7.5, 10 mM Na2S, 10 mM OAS, and 5 mM DTT. SAT activity was assayed by coupling to the OAS-TL reaction (Nakamura et al., 1987). To ensure a high excess of OAS-TL activity during coupling of both reactions, all SAT activity determinations were supplemented with 4 units of purified recombinant OAS-TL (Wirtz et al., 2004). Protein extracts from leaves were separated according to Laemmli (1970) by discontinuous SDS-PAGE in Mini-Protean II cells (Bio-Rad). Immunological detection of proteins was performed as described by Wirtz and Hell (2007), with the only difference that 20 μg of total proteins was blotted on nitrocellulose membranes. The dilution of purified primary antibodies for At SAT3 and At OAS-TL C was 1:10,000 and 1:1000, respectively.
Purification of the OAS-TL Protein Family
Aliquots of 100 g of leaf material from 9-week-old soil-grown Arabidopsis plants were ground in liquid nitrogen. For extraction of proteins, leaf powder was stirred for 30 min in 350 mL of ice-cold extraction buffer B (50 mM Tris-HCl, pH 8.0, 250 mM NaCl, 80 mM imidazole, 0.5 mM PMSF, and 1 mM DTT). The crude plant extract was filtered through a Miracloth tissue (Calbiochem), and the residual cell debris was collected by centrifugation for 20 min at 27,500 g and 4°C. The resulting supernatant was fractionated by successive precipitation of proteins with 20 and 75% ammonium sulfate on ice for 30 min, followed by centrifugation as described above to collect the proteins. The sediment was dissolved in 12 mL of extraction buffer A and desalted by size-exclusion chromatography using PD-10 columns in the same buffer (GE Healthcare) according to the manufacturer's instructions. The resulting protein solution was used for the purification of OAS-TL proteins by their interaction with recombinant At SAT5 (At5g56760). Recombinant At SAT5 fused to a hexa-His tag was expressed in Escherichia coli HMS174 (DE3) and immobilized on a nickel-loaded HiTrap column (GE Healthcare) according to the manufacturer's instructions (see Supplemental Figure 2 online). Extraction and binding buffer for the recombinant At SAT5 protein was 50 mM Tris-HCl, pH 8.0, 250 mM NaCl, 20 mM imidazole, and 0.5 mM PMSF. The immobilized At SAT5 was washed with 10 mL of washing buffer (50 mM Tris-HCl, pH 8.0, 250 mM NaCl, 80 mM imidazole, and 0.5 mM PMSF), and potential contamination with bacterial OAS-TL (CysK) was removed with 10 mL of elution buffer (50 mM Tris-HCl, pH 8.0, 250 mM NaCl, 80 mM imidazole, and 10 mM OAS). Elution of bacterial OAS-TL was the result of dissociation of the heterologous CSC by OAS. For that reason, OAS was removed from the column by washing with 10 mL of washing buffer prior to binding of plant OAS-TLs. Afterward, the plant protein solution was circulated over the column for 1 h in order to bind plant OAS-TLs to the recombinant At SAT5 protein. After washing with 10 mL of washing buffer, the plant OAS-TL proteins were eluted from the column with 10 mL of elution buffer and collected in 0.5-mL fractions. During the entire purification at room temperature, the flow rate was adjusted to 1 mL/min.
Separation of Purified OAS-TL Proteins by Two-Dimensional PAGE
Eighty micrograms of OAS-TL purified from wild-type and oastlC plants, 35 μg of OAS-TL purified from oastlA and oastlB plants, and 16 μg purified from oastlAB plants were precipitated for 2 h at −20°C with 1 mL of precipitation solution (acetone containing 10% [w/v] trichloroacetic acid and 0.07% β-mercaptoethanol). After centrifugation for 1 h at 25,000 g and 4°C, the supernatant was removed and the collected protein was resuspended in 1 mL of acetone containing 0.07% β-mercaptoethanol. The protein was recollected by centrifugation as described before. To minimize trichloroacetic acid contamination, this wash step was repeated twice. The resulting protein sediment was extensively dried and dissolved in 0.35 mL of rehydration solution (8 M urea, 2% [w/v] CHAPS, 0.005% [w/v] bromophenol blue, 0.2% [w/v] ampholytes pH 5 to 8 [Pharmacia], and 20 mM DTT). This solution was used to rehydrate a 17-cm IPG strip (Bio-Rad), pH 5 to 8, for 14 h with application of 50 V in the Protean IEF Cell (Bio-Rad). Following rehydration, the proteins were separated according to their isoelectric points with 4250 V for 20 h. After the isoelectric focusing, the IPG strip was equilibrated with SDS equilibration buffer (50 mM Tris-HCl, pH 8.8, 6 M urea, 30% [v/v] glycerin, 2% [v/v] SDS, 0.0025% [w/v] bromophenol blue, and 65 mM DTT) and placed on top of an 11% SDS gel. SDS-PAGE was performed as described above, but with Protean II xi cells (Bio-Rad).
Identification of Proteins by Mass Spectrometric Analyses
Protein species from two-dimensional PAGE were identified after in-gel digestion by MALDI-TOF-MS analysis combined with database searches as described by Colasante et al. (2006) and Melchers et al. (2007). The intact masses of the proteins were determined by electrospray ionization mass spectrometry analysis of the purified OAS-TL fractions according to Melchers et al. (2007).
Determination of Incorporation Rates Using 35SO42−
Twelve hours prior to the labeling experiment, hydroponically grown plants were enclosed in transparent plastic bags to ensure opening of the stomata. Leaf pieces (30 mg) were cut out from the interveinal fields of leaves and floated for 15 min on the labeling solution (half-strength Hoagland medium as above [with a total of 0.515 mM sulfate] containing 125 nM H235SO4 [1.5 mCi/nmol; Hartmann Analytic]) on a horizontal shaker with 60 rpm in the light (17 μE). The leaf pieces were washed twice with half-strength Hoagland medium and incubated for another 30 min on half-strength Hoagland medium while shaking at the same conditions as described above. Samples were taken after 5, 15, and 45 min, washed twice with half-strength Hoagland medium, and frozen in liquid nitrogen. A negative control for contamination of the leaf surface with 35SO42− was taken by dipping leaf pieces into the labeling solution for 1 s, prior to washing and harvesting like the samples. The samples were powdered using a Bio101 ThermoSavant Fast Prep system (Qbiogene) according to the manufacturer's instructions. The resulting powder was extracted and analyzed for thiols as described above. Eluent fractions containing Cys and GSH were collected independently and transferred in 10 mL of Ultima Gold scintillation amplifier (Perkin-Elmer). The incorporated radioactivity of the samples was determined for 10 min using standard settings for 35S in the liquid scintillation counter LS6000SC (Beckman-Coulter). For quantification of radioactively labeled sulfate, 50 μL of metabolite extract was mixed with 50 μL of 1 M BaCl2. The precipitated barium sulfate was collected by centrifugation for 10 min at 16,000 g and room temperature. The resulting sediment was dissolved in 0.8 mL of ammonium-EDTA solution (15.7 mM EDTA in a 0.75% ammonium hydroxide solution) and mixed with 10 mL of Ultima Gold scintillation amplifier for determination of the radioactivity in the liquid scintillation counter. Values obtained for the negative control were subtracted from the samples in order to exclude the radioactive sulfate from the leaf surface that had not been taken up. For quantification of radioactively labeled protein, the protein-containing pellet that was separated during HCl extraction was washed with 1 mL of 0.1 M HCl, resuspended in 1 mL of 8 M urea, and incubated overnight at 37°C. After centrifugation, 50 μL of the supernatant was precipitated using the 2-D Quant Kit (Amersham Biosciences) according to the manufacturer's instructions. The protein was dissolved and mixed with 8 mL of Ultima Gold scintillation amplifier for determination of radiolabel in the liquid scintillation counter.
Preparation of Mitochondrial Protein Extracts
Mitochondria were isolated from a heterotrophically grown Arabidopsis cell culture as described by Kruft et al. (2001). Proteins of isolated mitochondria were extracted in ME buffer (0.1 M Tris, pH 8.0, 50 mM NaCl, 2 mM DTT, and 1 mM PMSF) by three ultrasonic pulses with 40% energy for 10 s (Sonoplus GM 70; Bandelin Electronic). Cell debris was collected by centrifugation at 22,000 g and 4°C for 15 min. The resulting supernatant was used for the determination of SAT and OAS-TL activity.
Statistical Analysis
Comparison of means from different data sets was analyzed for statistical significance with the unpaired t test. Constant variance and normal distribution of data were checked with SigmaStat 3.0 prior to statistical analysis. The Mann-Whitney rank sum test was used to analyze samples that did not follow normal Gaussian distribution. Significance limits are indicated by asterisks: P < 0.05 > 0.01*; P < 0.01 > 0.001**; P < 0.001***.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At SAT3 (At3g13110), At SAT5 (NM_125059), At SIR (At5g04590), OASTLA1 (At4g14880), OASTLA2 (At3g22460), OASTLB (At2g43750), OASTLC (At3g59760), At CysD1 (At3g04940), At CysD2 (At5g28020), At CysC1 (At3g61440), At CS26 (At3g03630), and At CS-like (At5g28030).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Genomic Analysis of oastl T-DNA Insertion Lines.
Supplemental Figure 2. SAT Affinity Purification of Plant OAS-TL Proteins.
Supplemental Figure 3. Transcriptional Expression Pattern of CS26, CysC1, and CS-Like in the oastl Mutants A, B, C, and AB Compared with the Wild-Type Col-0 Determined by Quantitative PCR.
Supplemental Figure 4. Carbon, Nitrogen, Sulfur, and Nitrate Levels in Leaves of the Wild Type and oastl T-DNA Insertion Lines.
Supplementary Material
Acknowledgments
We gratefully acknowledge funding of C.H. by the Landesgraduiertenförderung Baden-Württemberg (BioQant Graduate School Molecular Machines) and of C.K. by the Olympia-Morata Program of the University of Heidelberg, the Schmeil-Stiftung, Heidelberg, and the Deutsche Forschungsgemeinschaft (Grant He1848/5-2). The kind provision of isolated mitochondria by H.-P. Braun is gratefully acknowledged. We thank S. Hassel for excellent technical assistance and A. Meyer for critically reading the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Rüdiger Hell (rhell@hip.uni-heidelberg.de).
Online version contains Web-only data.
References
- Allam, A.I., and Hollis, J.P. (1972). Sulfide inhibition in rice roots. Phytopathology 62 634–639. [Google Scholar]
- Berkowitz, O., Wirtz, M., Wolf, A., Kuhlmann, J., and Hell, R. (2002). Use of biomolecular interaction analysis to elucidate the regulatory mechanism of the cysteine synthase complex from Arabidopsis thaliana. J. Biol. Chem. 277 30629–30634. [DOI] [PubMed] [Google Scholar]
- Bogdanova, N., Bork, C., and Hell, R. (1995). Cysteine biosynthesis in plants: Isolation and functional identification of a cDNA encoding a serine acetyltransferase from Arabidopsis thaliana. FEBS Lett. 358 43–47. [DOI] [PubMed] [Google Scholar]
- Bogdanova, N., and Hell, R. (1997). Cysteine synthesis in plants: Protein-protein interactions of serine acetyltransferase from Arabidopsis thaliana. Plant J. 11 251–262. [DOI] [PubMed] [Google Scholar]
- Bonner, E.R., Cahoon, R.E., Knapke, S.M., and Jez, J.M. (2005). Molecular basis of cysteine biosynthesis in plants: Structural and functional analysis of O-acetylserine sulfhydrylase from Arabidopsis thaliana. J. Biol. Chem. 280 38803–38813. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Brunold, C., and Suter, M. (1982). Intracellular localization of serine acetyltransferase in spinach leaves. Planta 155 321–327. [DOI] [PubMed] [Google Scholar]
- Campisi, L., Yang, Y., Yi, Y., Heilig, E., Herman, B., Cassista, A.J., Allen, D.W., Xiang, H., and Jack, T. (1999). Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 17 699–707. [DOI] [PubMed] [Google Scholar]
- Colasante, C., Ellis, M., Ruppert, T., and Voncken, F. (2006). Comparative proteomics of glycosomes from bloodstream form and procyclic culture form Trypanosoma brucei brucei. Proteomics 6 3275–3293. [DOI] [PubMed] [Google Scholar]
- De Kok, L.J., Stuiver, C.E.E., Rubinigg, M., Westerman, S., and Grill, D. (1997). Impact of atmospheric sulfur deposition on sulfur metabolism in plants: H2S as sulfur source for sulfur deprived Brassica oleracea L. Bot. Acta 110 411–419. [Google Scholar]
- Dellaporta, S.L., Wood, J., Hicks, J. B. (1982). A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1 19–21. [Google Scholar]
- Droux, M. (2003). Plant serine acetyltransferase: New insights for regulation of sulphur metabolism in plant cells. Plant Physiol. Biochem. 41 619–627. [Google Scholar]
- Droux, M. (2004). Sulfur assimilation and the role of sulfur in plant metabolism: A survey. Photosynth. Res. 79 331–348. [DOI] [PubMed] [Google Scholar]
- Droux, M., Ruffet, M.L., Douce, R., and Job, D. (1998). Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants—Structural and kinetic properties of the free and bound enzymes. Eur. J. Biochem. 255 235–245. [DOI] [PubMed] [Google Scholar]
- Düring-Olsen, L., Regenberg, B., Gjermansen, C., Kielland-Brandt, M.C., and Hansen, J. (1999). Cysteine uptake by Saccharomyces cerevisiae is accomplished by multiple permeases. Curr. Genet. 35 609–617. [DOI] [PubMed] [Google Scholar]
- Fischer, W.N., Loo, D.D., Koch, W., Ludewig, U., Boorer, K.J., Tegeder, M., Rentsch, D., Wright, E.M., and Frommer, W.B. (2002). Low and high affinity amino acid H+-cotransporters for cellular import of neutral and charged amino acids. Plant J. 29 717–731. [DOI] [PubMed] [Google Scholar]
- Francois, J.A., Kumaran, S., and Jez, J.M. (2006). Structural basis for interaction of O-acetylserine sulfhydrylase and serine acetyltransferase in the Arabidopsis cysteine synthase complex. Plant Cell 18 3647–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitonde, M.K. (1967). A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occuring amino acids. Biochem. J. 104 627–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzam, A., Martin, M., Hell, R., and Meyer, A. (2007). γ-Glutamyl transpeptidase GGT4 initiates vacuolar degradation of glutathione S-conjugates in Arabidopsis. FEBS Lett. 581 3131–3138. [DOI] [PubMed] [Google Scholar]
- Gutensohn, M., et al. (2004). Characterization of a T-DNA insertion mutant for the protein import receptor atToc33 from chloroplasts. Mol. Genet. Genomics 272 379–396. [DOI] [PubMed] [Google Scholar]
- Hatzfeld, Y., Maruyama, A., Schmidt, A., Noji, M., Ishizawa, K., and Saito, K. (2000). β-Cyanoalanine synthase is a mitochondrial cysteine synthase-like protein in spinach and Arabidopsis. Plant Physiol. 123 1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell, R. (1997). Molecular physiology of plant sulfur metabolism. Planta 202 138–148. [DOI] [PubMed] [Google Scholar]
- Hell, R., Bork, C., Bogdanova, N., Frolov, I., and Hauschild, R. (1994). Isolation and characterization of two cDNAs encoding for compartment specific isoforms of O-acetylserine (thiol) lyase from Arabidopsis thaliana. FEBS Lett. 351 257–262. [DOI] [PubMed] [Google Scholar]
- Hell, R., and Hillebrand, H. (2001). Plant concepts for mineral acquisition and allocation. Curr. Opin. Biotechnol. 12 161–168. [DOI] [PubMed] [Google Scholar]
- Hell, R., Jost, R., Berkowitz, O., and Wirtz, M. (2002). Molecular and biochemical analysis of the enzymes of cysteine biosynthesis in the plant Arabidopsis thaliana. Amino Acids 22 245–257. [DOI] [PubMed] [Google Scholar]
- Hesse, H., Lipke, J., Altmann, T., and Hofgen, R. (1999). Molecular cloning and expression analyses of mitochondrial and plastidic isoforms of cysteine synthase (O-acetylserine(thiol)lyase) from Arabidopsis thaliana. Amino Acids 16 113–131. [DOI] [PubMed] [Google Scholar]
- Hirai, M., Fujiwara, T., Awazuhara, M., Kimura, T., Noji, M., and Saito, K. (2003). Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J. 33 651–663. [DOI] [PubMed] [Google Scholar]
- Jacques, A.G. (1936). The kinetics of penetration. XII. Hydrogen sulfide. J. Gen. Physiol. 19 397–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost, R., Berkowitz, O., Wirtz, M., Hopkins, L., Hawkesford, M.J., and Hell, R. (2000). Genomic and functional characterization of the oas gene family encoding O-acetylserine (thiol) lyases, enzymes catalyzing the final step in cysteine biosynthesis in Arabidopsis thaliana. Gene 253 237–247. [DOI] [PubMed] [Google Scholar]
- Kawashima, C.G., Berkowitz, O., Hell, R., Noji, M., and Saito, K. (2005). Characterization and expression analysis of a serine acetyltransferase gene family involved in a key step of the sulfur assimilation pathway in Arabidopsis. Plant Physiol. 137 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva, S. (2006). Regulation of sulfate assimilation in Arabidopsis and beyond. Ann. Bot. (Lond.) 97 479–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kredich, N.M., Becker, M.A., and Tomkins, G.M. (1969). Purification and characterization of cysteine synthetase, a bifunctional protein complex, from Salmonella typhimurium. J. Biol. Chem. 244 2428–2439. [PubMed] [Google Scholar]
- Kruft, V., Eubel, H., Jansch, L., Werhahn, W., and Braun, H.-P. (2001). Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 127 1694–1710. [PMC free article] [PubMed] [Google Scholar]
- Kuske, C.R., Hill, K.K., Guzman, E., and Jackson, P.J. (1996). Subcellular location of O-acetylserine sulfhydrylase isoenzymes in cell cultures and plant tissues of Datura innoxia Mill. Plant Physiol. 112 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuske, C.R., Ticknor, L.O., Guzman, E., Gurley, L.R., Valdez, J.G., Thompson, M.E., and Jackson, P.J. (1994). Purification and characterization of O-acetylserine sulfhydrylase isoenzymes from Datura innoxia. J. Biol. Chem. 269 6223–6232. [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Leustek, T., Martin, M.N., Bick, J.-A., and Davies, J.P. (2000). Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 141–165. [DOI] [PubMed] [Google Scholar]
- Leustek, T., and Saito, K. (1999). Sulfate transport and assimilation in plants. Plant Physiol. 120 637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn, J.E., Droux, M., Martin, J., and Douce, R. (1990). Localization of ATP-sulfurylase and O-acetylserine(thiol)lyase in spinach leaves. Plant Physiol. 94 1345–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas, F.M., van Loo, E.N., and van Hasselt, P.R. (1988). Effect of long-term H2S fumigation on photosynthesis in spinach. Correlation between CO2 fixation and chlorophyll fluorescence. Physiol. Plant. 72 77–83. [Google Scholar]
- Martin, M.N., Saladores, P.H., Lambert, E., Hudson, A.O., and Leustek, T. (2007). Localization of members of the gamma-glutamyl transpeptidase family identifies sites of glutathione and glutathione S-conjugate hydrolysis. Plant Physiol. 144 1715–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchers, J., Dirdjaja, N., Ruppert, T., and Krauth-Siegel, R.L. (2007). Glutathionylation of trypanosomal thiol redox proteins. J. Biol. Chem. 282 8678–8694. [DOI] [PubMed] [Google Scholar]
- Mino, K., Imamura, K., Sakiyama, T., Eisaki, N., Matsuyama, A., and Nakanishi, K. (2001). Increase in the stability of serine acetyltransferase from Escherichia coli against cold inactivation and proteolysis by forming a bienzyme complex. Biosci. Biotechnol. Biochem. 65 865–874. [DOI] [PubMed] [Google Scholar]
- Miranda, M., Borisjuk, L., Tewes, A., Heim, U., Sauer, N., Wobus, U., and Weber, H. (2001). Amino acid permeases in developing seeds of Vicia faba L.: Expression precedes storage protein synthesis and is regulated by amino acid supply. Plant J. 28 61–71. [DOI] [PubMed] [Google Scholar]
- Nakamura, K., Hayama, A., Masada, M., Fukushima, K., and Tamura, G. (1987). Measurement of serine acetyltransferase activity in crude plant extracts by a coupled assay system using cysteine synthase. Plant Cell Physiol. 28 885–891. [Google Scholar]
- Noctor, G., Gomez, L., Vanacker, H., and Foyer, C.H. (2002). Interactions between biosynthesis, compartmentation and transport in the control of glutathione homeostasis and signalling. J. Exp. Bot. 53 1283–1304. [DOI] [PubMed] [Google Scholar]
- Noji, M., Inoue, K., Kimura, N., Gouda, A., and Saito, K. (1998). Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase involved in cysteine biosynthesis from Arabidopsis thaliana. J. Biol. Chem. 273 32739–32745. [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu, N., Radwan, S., Peterson, A., Zhao, P., Badr, A., Xiang, C., and Oliver, D. (2007. a). Characterization of the extracellular gamma-glutamyl transpeptidases, GGT1 and GGT2, in Arabidopsis. Plant J. 49 865–877. [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu, N., Zhao, P., Xiang, C., and Oliver, D. (2007. b). Glutathione conjugates in the vacuole are degraded by gamma-glutamyl transpeptidase GGT3 in Arabidopsis. Plant J. 49 878–888. [DOI] [PubMed] [Google Scholar]
- Pasternak, M., Lim, B., Wirtz, M., Hell, R., Cobbett, C.S., and Meyer, A.J. (2008). Restricting glutathione biosynthesis to the cytosol is sufficient for normal plant development. Plant J. http://dx.doi.org/10.1111/j.1365–313.2007.03389.x. [DOI] [PubMed]
- Rabeh, W.M., and Cook, P.F. (2004). Structure and mechanism of O-acetylserine sulfhydrylase. J. Biol. Chem. 279 26803–26806. [DOI] [PubMed] [Google Scholar]
- Rebeille, F., Jabrin, S., Bligny, R., Loizeau, K., Gambonnet, B., Van Wilder, V., Douce, R., and Ravanel, S. (2006). Methionine catabolism in Arabidopsis cells is initiated by a γ-cleavage process and leads to S-methylcysteine and isoleucine syntheses. Proc. Natl. Acad. Sci. USA 103 15687–15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders, J., Zahedi, R.P., Pfanner, N., Meisinger, C., and Sickmann, A. (2006). Toward the complete yeast mitochondrial proteome: Multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 5 1543–1554. [DOI] [PubMed] [Google Scholar]
- Riemenschneider, A., Riedel, K., Hoefgen, R., Papenbrock, J., and Hesse, H. (2005). Impact of reduced O-acetylserine(thiol)lyase isoform contents on potato plant metabolism. Plant Physiol. 137 892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffet, M.L., Lebrun, M., Droux, M., and Douce, R. (1995). Subcellular distribution of serine acetyltransferase from Pisum sativum and characterization of an Arabidopsis thaliana putative cytosolic isoform. Eur. J. Biochem. 227 500–509. [DOI] [PubMed] [Google Scholar]
- Saito, K. (2004). Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol. 136 2443–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sirko, A., Blaszczyk, A., and Liszewska, F. (2004). Overproduction of SAT and/or OASTL in transgenic plants: A survey of effects. J. Exp. Bot. 55 1881–1888. [DOI] [PubMed] [Google Scholar]
- Smith, I.K. (1972). Studies of l-cysteine biosynthetic enzymes in Phaseolus vulgaris L. Plant Physiol. 50 477–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storozhenko, S., Belles-Boix, E., Babiychuk, E., Herouart, D., Davey, M.W., Slooten, L., Van Montagu, M., Inze, D., and Kushnir, S. (2002). γ-Glutamyl transpeptidase in transgenic tobacco plants. Cellular localization, processing, and biochemical properties. Plant Physiol. 128 1109–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, D., Kuras, L., Cherest, H., Blaiseau, P.L., and Surdin-Kerjan, Y. (1997). Regulation of gene expression in yeast sulphur metabolism. In Sulphur Metabolism in Higher Plants, W.J. Cram, L.J. De Kok, I. Stulen, C. Brunold, and H. Rennenberg, eds (Leiden, The Netherlands: Backhuys Publishers), pp. 27–37.
- Tocquin, P., Corbesier, L., Havelange, A., Pieltain, A., Kurtem, E., Bernier, G., and Perilleux, C. (2003). A novel high efficiency, low maintenance, hydroponic system for synchronous growth and flowering of Arabidopsis thaliana. BMC Plant Biol. 3 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter, A., Wolf, S., Steininger, H., Bogs, J., and Rausch, T. (2005). Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: Implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. Plant J. 41 15–30. [DOI] [PubMed] [Google Scholar]
- Warrilow, A.G., and Hawkesford, M.J. (2000). Cysteine synthase (O-acetylserine(thiol)lyase) substrate specificities classify the mitochondrial isoform as a cyanoalanine synthase. J. Exp. Bot. 51 985–993. [DOI] [PubMed] [Google Scholar]
- Wirtz, M., Berkowitz, O., Droux, M., and Hell, R. (2001). The cysteine synthase complex from plants. Mitochondrial serine acetyltransferase from Arabidopsis thaliana carries a bifunctional domain for catalysis and protein-protein interaction. Eur. J. Biochem. 268 686–693. [DOI] [PubMed] [Google Scholar]
- Wirtz, M., and Droux, M. (2005). Synthesis of the sulfur amino acids: Cysteine and methionine. Photosynth. Res. 86 345–362. [DOI] [PubMed] [Google Scholar]
- Wirtz, M., Droux, M., and Hell, R. (2004). O-Acetylserine(thiol)lyase: An enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana. J. Exp. Bot. 55 1785–1798. [DOI] [PubMed] [Google Scholar]
- Wirtz, M., and Hell, R. (2003). Production of cysteine for bacterial and plant biotechnology: Application of cysteine feedback-insensitive isoforms of serine acetyltransferase. Amino Acids 24 195–203. [DOI] [PubMed] [Google Scholar]
- Wirtz, M., and Hell, R. (2006). Functional analysis of the cysteine synthase protein complex from plants: Structural, biochemical and regulatory properties. J. Plant Physiol. 163 273–286. [DOI] [PubMed] [Google Scholar]
- Wirtz, M., and Hell, R. (2007). Dominant-negative modification reveals the regulatory function of the multimeric cysteine synthase protein complex in transgenic tobacco. Plant Cell 19 625–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, T., Zhu, X., and Masada, M. (1998). Purification and characterization of a novel cysteine synthase isozyme from spinach hydrated seeds. Biosci. Biotechnol. Biochem. 62 501–507. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, Y., Nakamura, T., Kusano, T., and Sano, H. (2000). Three Arabidopsis genes encoding proteins with differential activities for cysteine synthase and β-cyanoalanine synthase. Plant Cell Physiol. 41 465–476. [DOI] [PubMed] [Google Scholar]
- Zhao, C., Kumada, Y., Imanaka, H., Imamura, K., and Nakanishi, K. (2006). Cloning, overexpression, purification, and characterization of O-acetylserine sulfhydrylase-B from Escherichia coli. Protein Expr. Purif. 47 607–613. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.