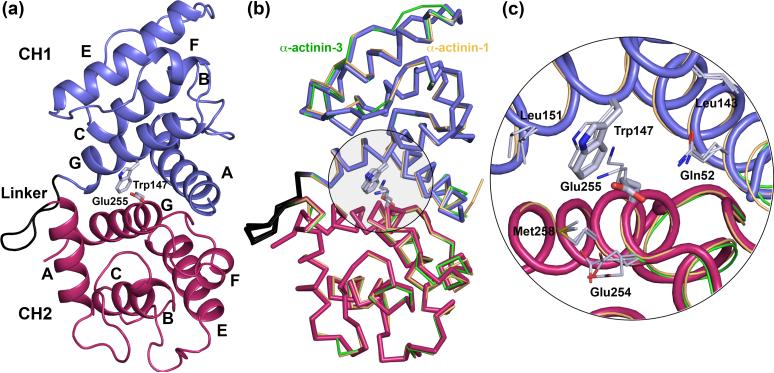
Figure 1.
Structure of the actin-binding domain (ABD) of α-actinin-4 Lys255Glu mutant. (a) Overall view of the structure of the ABD (CH1, blue; CH2, red). The linker loop between CH1 and CH2 (α-actinin-4 amino acids Asp157 to Ser164) is shown in black. Helices are labeled A to G (this ABD does not contain helix D). Note that although there are two molecules in the asymmetric unit of the crystal, their overall structures are very similar (Cα rms deviation of 0.5 Å) and only one molecule is shown (chain A). In addition to the amino acids of the ABD, chain A includes an extra Phe residue at the N-terminus (Phe46) and a Leu residue at the C-terminus (Leu272) from the expression plasmid. The side chains of Trp147 and the mutated amino acid Glu255 are shown. Trp147 and the wild type residue Lys255 are highly conserved in the spectrin family, making part of the actin-binding interface, and are typically involved in strong inter-CH contacts 14 (b) Superimpositions of the structures of the ABD of α-actinin-4 with those of α-actinin-1 (gold, Cα rms deviation 0.71 Å) and α-actinin-3 (green, Cα rms deviation 0.57 Å). Note that despite the presence of the Lys255Glu mutation in the ABD of α-actinin-4, the overall structures of the three α-actinin ABDs are nearly identical. (c) Close view of the Trp-Lys stacking interaction at the interface of CH1-CH2 in the ABDs of α-actinin-1 and α-actinin-3, and the Lys255Glu mutation in the ABD of α-actinin-4 that brakes this important interaction.
Experimental conditions: Fragments encoding the ABD of human α-actinin-4 (O43707, amino acids Ala47-Ala271) were generated by PCR from wild type and mutant α-actinin-4 cDNAs, subcloned into pGEX-4T1-HTa and verified by sequence analysis. GST-proteins were expressed in E. coli 5-alpha (NEB) and purified using Glutathione-Sepharose (GE Healthcare). Cleavage of the GST tag was carried out using Tobacco-Etch-Virus (TEV) protease, and proteins were purified using Ni Sepharose 6 Fast Flow (GE Healthcare). Protein purity and concentration were verified using SDS-PAGE and Bradford methods. The ABD of α-actinin-4 Lys255Glu mutant was concentrated to ∼8 mg ml−1 using Amicon Ultra centrifugal filters (Amicon) in 50 mM Tris-HCl pH 8.0, 100 mM NaCl and 1 mM DTT. Crystals were obtained using the hanging drop vapor diffusion method at 4°C by mixing 2 μl of the protein solution and 2 μl of a well solution containing 100 mM Imidazole pH 7.1, 50 mM NaCl, 1 mM EDTA, 5 % (v/v) glycerol, and 21 % (w/v) polyethylene glycol 5000 mono-methyl-ether. The percentage of glycerol was raised to ∼20 % for crystal freezing prior to X-ray data collection. An X-ray diffraction dataset was collected to the resolution of 2.2 Å from a crystal frozen in liquid nitrogen, and using the beamline F2 of the Cornell High Energy Synchrotron Source (CHESS). The diffraction data were indexed and scaled with the program HKL2000 (HKL Research, Inc) (Table 1). A molecular replacement solution for the two independent ABD molecules in the asymmetric unit of the crystals was obtained with the program AMoRe 36, and using the 1.7 Å resolution crystal structure of the ABD of α-actinin-1 14 as a search model. Model building and refinement were carried out with the program Coot 37, and the CCP4 program Refmac. The refinement of the structure converged to R-factor and R-free values of 17.1% and 22.4 %, respectively (Table 1). The electron density map reveals all the amino acids of the two ABD molecules in the asymmetric unit of the crystal. The construct crystallized here contained extra amino acids at the N-terminus (GAMDPEF) and the C-terminus (LDELN) resulting from the expression vector. Some of these amino acids are also observed in the electron density map and form part of the refined model (PDB Code 2R0O).