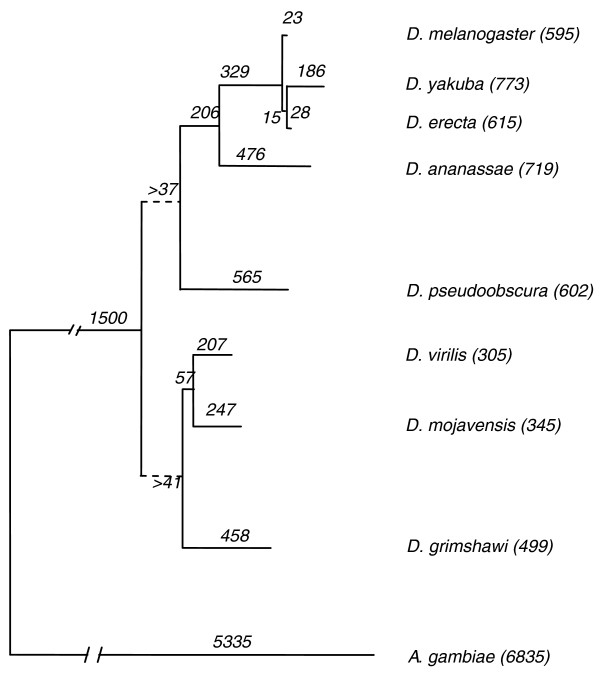
Figure 3.
Rearrangement phylogeny for genus Drosophila. The number along each branch of the tree shows the probable number of fixed rearrangement breaks inferred along that evolutionary branch. Each inferred rearrangement break corresponds to the disruption of a gene pair (NGP) that was inferred to exist in the immediate ancestor. Consequently, it includes macro and micro syntenic disruptions. See Materials and methods for details on the handling of ambiguous cases. Rearrangement breaks are assumed to occur as a result of chromosomal inversion events. Estimates for inversion counts can be computed from these data as outlined in the Materials and methods. The total number of inferred fixed rearrangement breaks for each genus Drosophila species, from the Drosophila root, is mentioned alongside the species name. Anopheles gambiae (shown), Aedes aegypti, Apis mellifera, and Tribolium castaneum are also used as outgroup species. Subgenus Drosophila species show lower overall average branch lengths than subgenus Sophophora species. Dashed lines at the subgenus Sophophora and subgenus Drosophila nodes reflect the loss of genus-specific NGP signal at the genus Drosophila root, which is only partially compensated for by distant outgroup species. See Discussion for details.