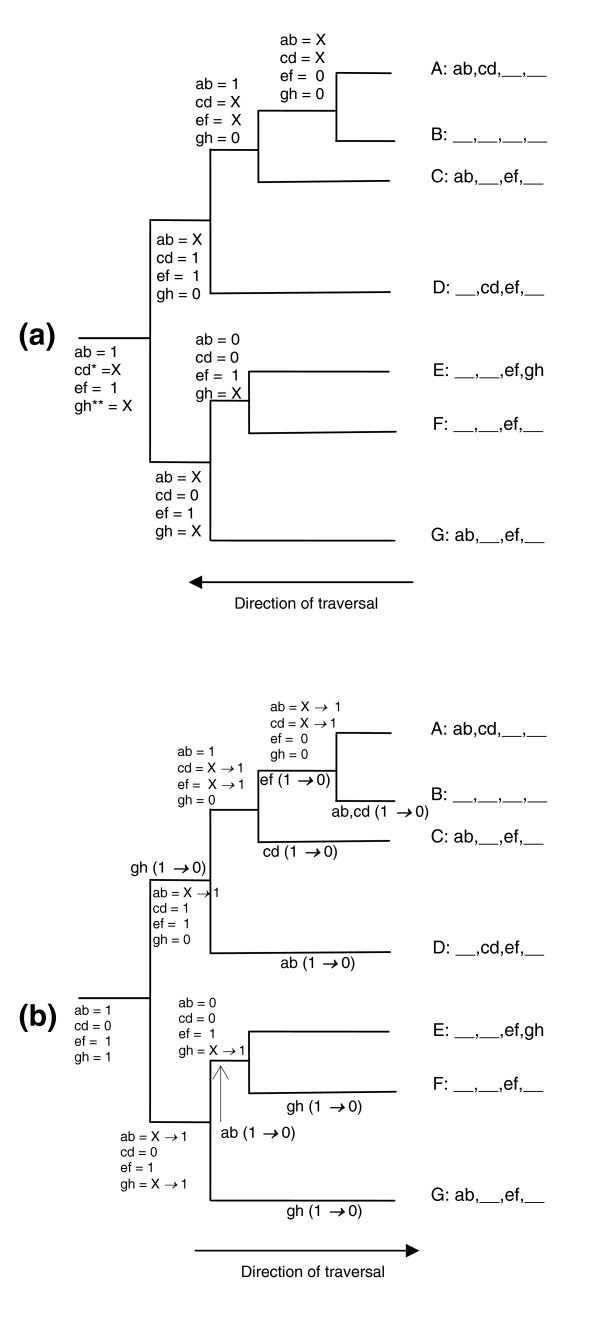
Figure 5.
Two-stage tree traversal algorithm example. Species A through G are shown with representative gene pair content (four pairs: ab, cd, ef, gh; an underscore '_' implies that that pair does not exist in that species). The state of pairs at each node is shown and state transitions are shown in bold font. (a) Leaf-to-root traversal. Ancestral states of gene pairs are assigned with the constraint that a gene can be in at most two pairs at any given node. A '1' implies that the pair exists at a given node where at least one species on either side of the node has that pair. A '0' implies that it does not exist in any leaf species reachable from that node. An 'X' implies that the state is unknown due to conflicting 1/0 or X/0 information from child nodes (that is, a '1'/'X' exists for that pair on one side of the node and a '0' on the other side). 0 → X, 1 → X, and X → 1 transitions are seen during this leaf-to-root tree traversal. In the case of pairs like cd*, where a '1' and '0' are inferred at the child nodes at the root of the tree, and there is no further evidence from outgroup species, the state is left undetermined and does not contribute to rearrangement analysis. It is hoped that addition of more genomes in this analysis will help resolve this in the future. In cases where the root value is 'X' (as in pair gh**), it is set to '1' if an outgroup species has this pair (given that it already exists in at least one non-outgroup species), else it does not contribute to this analysis. (b) Root-to-leaf traversal. Pair gh is assumed to be set to '1' at the root of the tree for this example, using the criteria above. Rearrangements are assigned to tree branches. A 0 → 1 transition reflects creation of a pair that did not exist at an ancestral state, including pairs unique to a species. A 1 → 0 transition represents a pair being lost due to a rearrangement. X → 0 and X → 1 transitions at nodes represent inheritance of an inferred ancestral state where the current value is unknown due to conflicting child evidence. The rearrangement phylogeny counts the number of 1 → 0 transitions (NGP disruptions) along each branch. See Additional data file 10 for a detailed description of the method.