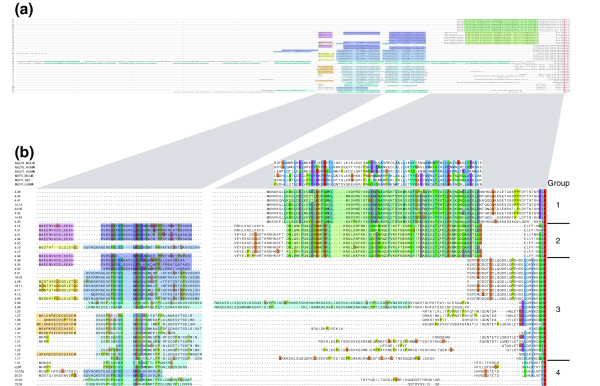
Figure 15.
Structure of the amino-termini of the new zebrafish NLR proteins. ClustalW alignment of the set of 70 predicted NLR proteins was truncated four amino acids downstream of the start of the Fisna domain, and the alignment of the remaining amino-terminal sequences was edited manually using Jalview [62]. Sequences that did not extend significantly beyond the Fisna domain were deleted, as were some sequences in groups with many similar or identical sequences. The remaining sequences represent a set of characteristic compositions of motifs found in the amino-terminal part of this family of proteins. (a) Overview of the alignment with characteristic sequence motifs shaded in color: green = pyrin-like domain in groups 1 and 2; blue = repeated motif (different shades of blue mark different versions of the repeat); yellow/orange tones = conserved amino-terminal amino acids; and pink = specific amino-terminal peptide of 14 amino acids. (b) Details of the alignment in panel a in which amino acid similarities and identities are highlighted in ClustalW colors. A set of mammalian PYD domains are aligned above the zebrafish group 1 and group 2 pyrin-like domains to illustrate the similarity. NLR, nucleotide-binding domain/NACHT domain and leucine rich repeat containing family.