Abstract
Stavudine is a hepatotoxic antiretroviral nucleoside analogue that also inhibits the replication of mitochondrial DNA (mtDNA). To elucidate the mechanism and consequences of mtDNA depletion, we treated HepG2 cells with stavudine and either redoxal, an inhibitor of de novo pyrimidine synthesis, or uridine, from which pyrimidine pools are salvaged. Compared with treatment with stavudine alone, co-treatment with redoxal accelerated mtDNA depletion, impaired cell division, and activated caspase 3. These adverse effects were completely abrogated by uridine. Intracellular ATP levels were unaffected. Transcriptosome profiling demonstrated that redoxal and stavudine acted synergistically to induce CDKN2A and p21, indicating cell cycle arrest in G1, as well as genes involved in intrinsic and extrinsic apoptosis. Moreover, redoxal and stavudine showed synergistic interaction in the up-regulation of transcripts encoded by mtDNA and the induction of nuclear transcripts participating in energy metabolism, mitochondrial biogenesis, oxidative stress, and DNA repair. Genes involved in nucleotide metabolism were also synergistically up-regulated by both agents; this effect was completely antagonized by uridine. Thus, pyrimidine depletion sensitizes cells to stavudine-mediated mtDNA depletion and enhances secondary cell toxicity. Our results indicate that drugs that diminish pyrimidine pools should be avoided in stavudine-treated human immunodeficiency virus patients. Uridine supplementation reverses this toxicity and, because of its good tolerability, has potential clinical value for the treatment of side effects associated with pyrimidine depletion.
Highly active antiretroviral therapy usually includes nucleoside analogue reverse-transcriptase inhibitors (NRTIs) in its backbone and has resulted in a significant decrease of human immunodeficiency virus (HIV)-associated morbidity and mortality. Several side effects have been associated with the long-term use of NRTIs and their ability to inhibit polymerase-γ, the human enzyme that replicates mitochondrial DNA (mtDNA).1 Decreased mtDNA copy numbers result in an impaired synthesis of mtDNA-encoded respiratory chain subunits and a secondary defect of oxidative phosphorylation.2,3 Many organs can be involved.1,4 The mitochondrial toxicity of NRTIs in the liver has been associated with steatosis, steatohepatitis, and acute organ failure and probably also contributes to the hyperlactatemia and lactic acidosis that continues to be a problem in resource-poor settings.5,6,7,8,9,10,11
By exposing hepatocytes and adipocytes to pyrimidine NRTI in vitro, we have recently discovered that uridine prevents and even reverses the mitochondrial toxicity of these antiretrovirals.12,13 In mice, a dietary supplement with a high bioavailability of uridine was shown to prevent zalcitabine-induced microvesicular steatohepatitis.6 In humans, uridine is well tolerated.14 Several clinical trials currently investigate the dietary supplementation of uridine in the prevention of NRTI-related side effects (http://www.clinicaltrials.gov). The first results are promising.15,16,17
The mechanism of the protective effects of uridine has not been fully delineated.14 Severe mtDNA depletion and secondary respiratory chain dysfunction is thought to diminish the availability of intracellular pyrimidines, because a normal electron flux through the respiratory chain is required for the activity of dihydroorotate dehydrogenase (DHODH), an enzyme that is essential for pyrimidine de novo synthesis.18 Intramitochondrial pyrimidine deficiency could then induce or contribute to an even more profound mtDNA-depletion by allowing the triphosphorylated pyrimidine antiretrovirals to compete more efficiently with their natural pyrimidine counterparts at polymerase-γ. Exogenous uridine supplementation may disrupt this vicious circle by replenishing intracellular pyrimidine pools distal from DHODH through the salvage pathway.14 Alternatively uridine (or its metabolites) may compete with antiretroviral nucleoside prodrugs either at carriers permitting their intramitochondrial import, or at enzymes responsible for their intracellular activation.19
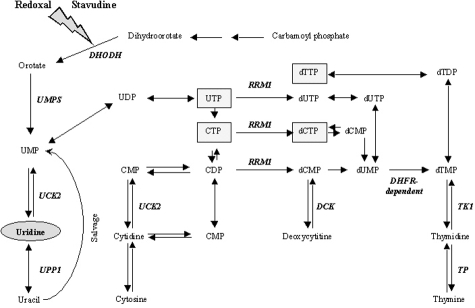
The aim of our study was to better characterize the mechanism of the beneficial effects of uridine on hepatocytes and to more specifically examine, if pyrimidine depletion does indeed contribute to the mitochondrial toxicity by sensitizing for the adverse effects of pyrimidine analogues. We therefore examined the toxic effects of stavudine in hepatocytes that were co-incubated with or without redoxal, an agent that induces pyrimidine depletion by directly and specifically blocking DHODH (Figure 1).20,21
Figure 1.
Metabolic pathways involved in pyrimidine metabolism. Uridine replenishes all pyrimidines distal from DHODH. Gray boxes indicate RNA and DNA building blocks. Based on the Kyoto Encyclopedia of Genes and Genomes21 reference pathway for pyrimidine metabolism (Accessed February 2007, http://www.genome.jp/kegg/pathway/map/map00240.html).
Materials and Methods
Materials
The human hepatoma HepG2 cell line was provided by the American Type Culture Collection, Rockville, MD (no. HB-8065). Cell culture flasks (75 cm2) were purchased from Becton Dickinson (San Jose, CA) and 10% fetal bovine serum from PAA Laboratories (Linz, Austria). Stavudine, redoxal, and uridine were purchased from Sigma (Taufkirchen, Germany).
Cell Culture
HepG2 cells were propagated at 37°C and 5% CO2 in Dulbecco’s modified Eagle medium, containing 4.5 g/L glucose and 110 mg/L pyruvate, supplemented with 10% fetal bovine serum, 50 U/ml streptomycin, 50 U/L penicillin, and 250 μg/L amphotericin B. HepG2 cells (2.7 × 106) were seeded during logarithmic growth at day 1 and harvested at days 5, 10, 15, 20, and 25. Cells were counted in a CASY cell counter (Schärfe System, Reutlingen, Germany) and 2.7 × 106 cells were replated in new flasks. Medium was renewed at the time of harvest and on every third day after plating.2
Stavudine was added in concentrations corresponding to the stavudine steady state peak plasma concentration of 3.6 μmol/L (Cmax) in humans during HIV therapy.2 Stavudine-exposed cells were also incubated with or without uridine (200 μmol/L) and redoxal, an inhibitor of DHODH.20 The concentration of redoxal (20 μmol/L) was chosen to slow down growth of HepG2 cells by ∼50%. Control HepG2 cells were cultured in medium without stavudine, uridine, or redoxal. A second set of HepG2 control cells was incubated with uridine alone.
MtDNA Copy Numbers
Total DNA was extracted with the QIAamp DNA isolation kit (Qiagen, Hilden, Germany). MtDNA and nDNA copy numbers were determined by quantitative polymerase chain reaction (PCR) using the ABI 7700 sequence detection system (Applied Biosystems, Foster City, CA). We amplified the mtDNA ATP-6 gene between nucleotide positions 8981 and 9061 with the forward primer, 5′-ACCAATAGCCCTGGCCGTAC-3′ and the backward primer 5′-GGTGGCGCTTCCAATTAGGT-3′. MtDNA was quantified with a FAM-fluorophore labeled probe (5′-6FAM-CCTAACCGCTAACATTACTGCAGGCCACC-TAMRA-3′). For the detection of nDNA we selected exon number 8 of the GAPDH gene between nucleotide positions 4280 and 4342, using the forward primer 5′-CGGGGCTCTCCAGAACATC-3′ and the backward primer 5′-ATGACCTTGCCCACAGCCT-3′. In this case we used a VIC-fluorophore-labeled probe (5′-VIC-CCCTGCCTCTACTGGCGCTGCC-TAMRA-3′).
Each 25-μl reaction contained 25 ng of genomic DNA, 100 nmol/L probe, 200 nmol/L primers, and TaqMan universal master mix (Applied Biosystems). Amplifications of mitochondrial and nuclear products were separately performed in optical 96-well plates (Applied Biosystems). An initial incubation at 50°C for 2 minutes was followed by 10 minutes at 95°C and 40 denaturing steps at 95°C (15 seconds), alternating with combined annealing/extension at 60°C (1 minute). All samples were run in triplicate. Absolute mtDNA and nDNA copy numbers were calculated using serial dilutions of plasmids with known copy numbers.22
Respiratory Chain Subunits
The subunit II of cytochrome c-oxidase (COXII) is encoded by mtDNA, whereas the subunit IV of cytochrome c-oxidase (COXIV) is encoded by nDNA. COXII was quantified by immunoblot from 106 HepG2 cells and normalized to the signal of a simultaneously used antibody against COXIV as described.22 Signal intensities were densitometrically quantified using Scion Image (Scion Corp., Frederick, MD).
Determination of Intracellular ATP
ATP was extracted from 106 cells and measured in a microplate luminometer (LB96V; Berthold-Technologies, Wildbad, Germany) against an ATP standard (10−4 mol/L to 10−12 mol/L) using an ATP bioluminescence assay kit (HSII; Roche, Penzberg, Germany).
Lipid Content and Lipid Peroxidation
Lipids were freshly extracted from 1 × 106 HepG2 cells using methanol/chloroform/water according to the Bligh-Dyer method and quantified spectrophotometrically by means of a sulfo-phospho-vanillin reaction on lipid standards (Sigma) as described.23 Malondialdehyde is one of the end products of lipid peroxidation and an indicator of free radical production and oxidative stress. Malondialdehyde was spectrophotometrically quantified in 1 × 106 HepG2 cells with an assay for thiobarbituric acid reactive material.24
Caspase 3 Activation
Activation of caspase 3 plays a central role in the execution of apoptotic cell death.25 Activated caspase 3 can be sensitively detected in living cells by staining with by Red-DEVD-FMK, a cell permeable, nontoxic dye (CaspGlow Red; MBL Laboratories Woburn, MA). Cells were stained with 1 μl of Red-DEVD-FMK per 300 μl of medium for 30 minutes at 37°C according to the manufacturer’s instruction. Red fluorescence was visualized using a spectral confocal microscope (Leica TCS SP2 AOBS; Leica Microsystems, Bensheim, Germany).
Real-Time Quantification of Nuclear and Mitochondrial Gene Transcripts
After 10 days of incubation, RNA was extracted from 1 × 106 cells (RNeasy kit, Qiagen). Quantity and integrity of the RNA were verified using RNA 6000 nano chips (2100 Bioanalyzer; Agilent, Palo Alto, CA). Five μg of RNA from each sample was reverse-transcribed using 200 U Superscript II (Invitrogen, Carlsbad, CA) and 100 μmol/L oligo(dT)12-18 (Invitrogen) as primer.
Primer pairs were designed with the aid of the universal probe library from Roche Diagnostics Corp. (Indianapolis, IN) in an intron-spanning manner to avoid unspecific amplification of contaminating genomic DNA. Primer sequences are shown in Table 1. Gene expression was quantified using the Light Cycler 489 (Roche) on a 384-well plate. Ten-μl reactions contained 5 μl of SYBR Green I master mix (Roche), 50 ng cDNA template, and 0.5 μmol/L of each primer. Target genes were run in duplicate on a single plate, which included the samples from all treatments, plus a no template control. Light cycling conditions were as follows: activation (95°C for 10 seconds), 40 amplification cycles (95°C for 10 seconds, 52°C for 5 seconds, and 72°C for 12 seconds). PCR conditions were optimized for all genes to ensure a PCR efficiency of 2. Melting curve analysis was done to ensure that all transcripts under investigation were represented by a single peak, indicating specificity. Gene expression was calculated from the real-time PCR efficiency26 in relation to the mean of four housekeeping genes (LMNB1, GAPDH, ACTB, and 36B4) commonly used.27,28 To estimate the reliability of our assays, proliferating cell nuclear antigen mRNA was measured in duplicate using two different primer sets. Proliferating cell nuclear antigen mRNA copies varied by a mean factor of 1.3 (SD, 0.34) between the two primer sets. For gene abbreviations, please refer to Table 2.
Table 1.
Primers Used in Real-Time Quantification of Nuclear and Mitochondrial Gene Transcripts
| Gene transcript | Forward | Reverse |
|---|---|---|
| Housekeeping | ||
| LMNB1 | 5′-AGCTGGAGTGGTTGTTGAGG-3′ | 5′-TTGGATGCTCTTGGGGTTC-3′ |
| GAPDH | 5′-CGGGCTCTCCAGAACATC-3′ | 5′-ATGACCTTGCCCACAGCCT-3′ |
| ACTB | 5′-ATTGGCAATGAGCGGTTC-3′ | 5′-GGATGCCACAGGACTCCAT-3′ |
| 36B4 | 5′-CTGGAAAACAACCCAGCTCT-3′ | 5′-GAACACAAAGCCCACATTCC-3′ |
| Cell cycle | ||
| PCNA | 5′-TGTCACAGACAAGTAATGTCGATAAA-3′ | 5′-GAACTGGTTCATTCATCTCTATGG-3′ |
| UbcH10 | 5′-CTGGAAAAACCCCACAGC-3′ | 5′-AAAAGACGACACAAGGACAGG-3′ |
| p21 | 5′-TCACTGTCTTGTACCCTTGTGC-3′ | 5′-GGCGTTTGGAGTGGTAGAAA-3′ |
| CDKN2A | 5′-TTCCCCCACTACCGTAAATG-3′ | 5′-CCAGAAAACTCCAACACAGTGA-3′ |
| Apoptosis | ||
| BAD | 5′-ACCAGCAGCAGCCATCAT-3′ | 5′-GGTAGGAGCTGTGGCGACT-3′ |
| BCL2 | 5′-AGGTGCATCTGGTGATGTGA-3′ | 5′-CACTCCAACCCCCGATCT-3′ |
| BCL2L1 | 5′-AACGTCATCCGCTACATCGT-3′ | 5′-CCCCAGAACCACAAAATCC-3′ |
| BAX | 5′-ATGTTTTCTGACGGCAACTTC-3′ | 5′-ATCAGTTCCGGCACCTTG-3′ |
| FAS | 5′-TGAATCTCCAACCTTAAATCCTG-3′ | 5′-TGACTCCAGCAATAGTGGTGAT-3′ |
| FASLG | 5′-CAGTCCACCCCCTGAAAA-3′ | 5′-GGACCTTGAGTTGGACTTGC-3′ |
| CASP3 | 5′-TGGAATTGATGCGTGATGTT-3′ | 5′-TGGCTCAGAAGCACACAAAC-3′ |
| CASP8 | 5′-TCCAAATGCAAACTGGATGA-3′ | 5′-CCCAGGATGACCCTCTTCTC-3′ |
| CASP9 | 5′-CCATATGATCGAGGACATCCA-3′ | 5′-GACTCCCTCGAGTCTCCAGAT-3′ |
| Mitochondrial energy metabolism | ||
| COX IV | 5′-CACCGCGCTCGTTATCAT-3′ | 5′-TGGCCACCCACTCTTTGT-3′ |
| ATP9 | 5′-TGCAGGGTAGTAGGAGTGCAG-3′ | 5′-TTAGACCCCTGGTACAACAGC-3′ |
| 16S rRNA | 5′-CGATTAAAGTCCTACGTGATCTGA-3′ | 5′-AGGGAGGAATTTGAAGTAGATAGAAA-3′ |
| ND1 | 5′-TCCACCCTTATCACAACACAAG-3′ | 5′-TCATATTATGGCCAAGGGTCA-3′ |
| COX I | 5′-GCTTCTGACTCTTACCTCCCTCT-3′ | 5′-CCGGCCTCCACTATAGCA-3′ |
| ATP6 | 5′-TTTATTGCCACAACTAACCTCCT-3′ | 5′-TTGGGTGGTTGGTGTAAATG-3′ |
| ND6 | 5′-GGTGCTGTGGGTGAAAGAGT-3′ | 5′-AACCCTGACCCCTCTCCTT-3′ |
| Mitochondrial biogenesis | ||
| POLG | 5′-GAGAAGGCCCAGCAGATGTA-3′ | 5′-ATCCGACAGCCGATACCA-3′ |
| PGC-1 | 5′-GAAGAGGAAGAAGGGGAGGA-3′ | 5′-GGTAGGGGCAGTGGTCAG-3′ |
| TFAM | 5′-GTTTCTCCGAAGCATGTGG-3′ | 5′-GCACAGCTCTGCTCCAGAC-3′ |
| Nucleotide metabolism | ||
| DHFR | 5′-TGCCTTTGAGAAATGAATGAAG-3′ | 5′-GCCCAAGCTGGTCTCAAGT-3′ |
| DGUOK | 5′-TCTCCATCGAAGGCAACATT-3’ | 5′-TGGAATGTGTAGGACCATCG-3′ |
| HPRT1 | 5′-TGACCTTGATTTATTTTGCATACC-3′ | 5′-CGAGCAAGACGTTCAGTCCT-3′ |
| DHODH | 5′-AGAAGCAGGCCAAGCTCAC-3′ | 5′-TGTTCTTCCCCAAGTTGACC-3′ |
| UMPS | 5′-AGGAAAGAAACAAAGGATTATGGA-3′ | 5′-TGGTGACAACATCTTCAATGATTA-3′ |
| RRM1 | 5′-GGCAAACTCACTAGTATGCACTTC-3′ | 5′-AAATAATACATCCCAGTCTTCAAACC-3′ |
| TP | 5′-ATCCAGAGCCCAGAGCAG-′ | 5′-CCAGCTGCTCACTCTGACC-3′ |
| UPP1 | 5′-TGGCACTTCTGGTGGGATA-3′ | 5′-CCACTGCCTGCTCTGTTATG-3′ |
| UCK2 | 5′-GATCATCCCTAGAGGTGCAGA-3′ | 5′-GGATGTCCTGGATGTGCTG-3′ |
| TK1 | 5′-CAGCTTCTGCACACATGACC-3′ | 5′-CGTCGATGCCTATGACAGC-3′ |
| DCK | 5′-AAATATGAAAGTCTGGTTGAAAAGG-3′ | 5′-AAAGCTGAAGTATCTGGAACCATT-3′ |
| Oxidative stress | ||
| GPX1 | 5′-ATGTGTGCTGCTCGGCTA-3′ | 5′-CGAGAAGGCATACACCGACT-3′ |
| SOD1 | 5′-TCATCAATTTCGAGCAGAAGG-3′ | 5′-CAGGCCTTCAGTCAGTCCTTT-3′ |
| SOD2 | 5′-GTGGTGGAGAACCCAAAGG-3′ | 5′-TGTCAAAGGAACCAAAGTCCA-3′ |
| DNA repair | ||
| UNG1 | 5′-TCGAATGGCCTTGTTTTCTT-3′ | 5′-TACATGGTGCCGCTTCCT-3′ |
| OGG1 | 5′-ATGGGGCATCGTACTCTAGC-3′ | 5′-AGGACTTTGCTCCCTCCAC-3′ |
| APE1 | 5′-TGGGGATAAAAGCCACTGC-3′ | 5′-CCAGCGAGTTTTCATCTGGT-3′ |
Table 2.
Transcriptional Regulation of Genes
| Uridine | Redoxal | Redoxal + uridine | Stavudine | Stavudine + uridine | Stavudine + redoxal | Stavudine + redoxal + uridine | |
|---|---|---|---|---|---|---|---|
| Cell cycle | |||||||
| PCNA | 1.2 ± 0.1 | 1.0 ± 0.1 | 0.8 ± 0.2 | 1.0 ± 0.1 | 0.8 ± 0.3 | 1.2 ± 0.1 | 1.1 ± 0.1 |
| UbcH10 | 1.2 ± 0.1* | 1.1 ± 0.1 | 1.0 ± 0.2 | 1.2 ± 0.1* | 0.8 ± 0.3 | 1.6 ± 0.1* | 1.4 ± 0.2* |
| p21 | 1.4 ± 0.1 | 2.7 ± 0.2** | 2.2 ± 0.4* | 2.1 ± 0.7* | 1.9 ± 0.1* | 4.2 ± 1.4* | 2.7 ± 0.2*** |
| CDKN2A | 1.2 ± 0.2 | 2.1 ± 0.6* | 1.3 ± 0.7 | 3.6 ± 0.9** | 1.9 ± 0.4*††† | 8.1 ± 3.3** | 2.2 ± 0.7*† |
| Apoptosis | |||||||
| BAD | 1.4 ± 0.1 | 2.7 ± 0.6* | 1.7 ± 0.4*† | 2.1 ± 1.0* | 1.8 ± 0.1* | 4.3 ± 1.8* | 2.7 ± 0.1*** |
| BCL2 | 1.5 ± 0.1 | 3.2 ± 0.1*** | 1.7 ± 0.3*††† | 2.0 ± 0.5* | 1.9 ± 0.2* | 4.3 ± 1.3* | 2.6 ± 0.2*** |
| BCL2L1 | 1.1 ± 0.1 | 2.7 ± 0.1*** | 0.9 ± 0.2 | 1.4 ± 0.1* | 1.1 ± 0.1† | 3.6 ± 0.1*** | 1.4 ± 0.3††† |
| BAX | 1.1 ± 0.1 | 1.4 ± 0.1* | 0.9 ± 0.2† | 1.1 ± 0.1 | 1.0 ± 0.1 | 1.9 ± 0.2* | 1.1 ± 0.1†† |
| FAS | 1.6 ± 0.4 | 4.9 ± 1.4* | 1.0 ± 0.1††† | 1.2 ± 0.2 | 1.4 ± 0.3 | 6.6 ± 0.3*** | 1.6 ± 0.1**††† |
| FASLG | 1.5 ± 0.2 | 3.5 ± 0.4** | 2.5 ± 0.7* | 2.5 ± 1.2 | 1.8 ± 0.1*† | 7.0 ± 2.1** | 3.8 ± 0.9** |
| CASP3 | 1.1 ± 0.1 | 2.0 ± 0.1** | 1.0 ± 0.2†† | 1.2 ± 0.1 | 1.1 ± 0.1 | 3.0 ± 0.1*** | 1.4 ± 0.1**††† |
| CASP8 | 1.2 ± 0.1 | 1.9 ± 0.1* | 1.0 ± 0.1†† | 1.4 ± 0.1* | 1.1 ± 0.1† | 2.5 ± 0.2** | 1.3 ± 0.2†† |
| CASP9 | 1.3 ± 0.1* | 2.2 ± 0.1*** | 0.8 ± 0.1††† | 1.3 ± 0.1* | 1.3 ± 0.1* | 3.6 ± 0.4** | 1.7 ± 0.2**†† |
| Mitochondrial energy metabolism | |||||||
| COX IV | 1.2 ± 0.1 | 2.1 ± 0.1** | 1.0 ± 0.2† | 1.2 ± 0.1 | 1.2 ± 0.1* | 2.8 ± 0.2** | 1.5 ± 0.2†† |
| ATP9 | 1.3 ± 0.1 | 3.1 ± 0.3** | 0.8 ± 0.4† | 1.4 ± 0.1* | 1.0 ± 0.2† | 4.3 ± 0.1*** | 1.7 ± 0.2**††† |
| 16S rRNA | 1.2 ± 0.4 | 2.5 ± 1.1* | 0.6 ± 0.1† | 4.1 ± 1.3** | 1.3 ± 0.7††† | 2.6 ± 0.6** | 1.2 ± 0.8* |
| ND1 | 0.8 ± 0.2 | 1.3 ± 0.2* | 0.9 ± 0.1 | 1.1 ± 0.3 | 0.6 ± 0.1* | 2.0 ± 0.6* | 1.0 ± 0.4* |
| COX I | 0.9 ± 0.1 | 1.3 ± 0.4 | 0.6 ± 0.3 | 0.8 ± 0.3 | 1.1 ± 0.2 | 2.3 ± 0.4* | 1.0 ± 0.2†† |
| ATP6 | 0.9 ± 0.1 | 1.6 ± 0.4 | 1.1 ± 0.2† | 0.8 ± 0.1 | 1.2 ± 0.6 | 2.9 ± 0.6** | 0.7 ± 0.1* |
| ND6 | 1.0 ± 0.1 | 2.7 ± 1.1* | 1.5 ± 0.2*† | 1.1 ± 0.1 | 1.6 ± 0.5 | 5.3 ± 2.4* | 1.2 ± 0.8*† |
| Mitochondrial biogenesis | |||||||
| POLG | 1.0 ± 0.1 | 1.7 ± 0.5 | 0.9 ± 0.1† | 1.1 ± 0.2 | 1.4 ± 0.5 | 2.3 ± 0.8* | 1.7 ± 0.8* |
| PGC-1 | 1.6 ± 0.1 | 3.0 ± 0.4** | 2.2 ± 0.3* | 2.4 ± 0.1* | 2.4 ± 0.1* | 5.3 ± 2.3* | 3.8 ± 0.3*** |
| TFAM | 1.7 ± 0.6 | 1.9 ± 0.6 | 1.2 ± 0.4† | 1.2 ± 0.2 | 1.2 ± 0.2 | 1.8 ± 0.6 | 1.4 ± 0.3 |
| Nucleotide metabolism | |||||||
| DHFR | 0.9 ± 0.1 | 1.9 ± 0.4* | 0.5 ± 0.2*† | 0.9 ± 0.1 | 0.9 ± 0.1 | 2.6 ± 0.6* | 1.0 ± 0.3*† |
| DGUOK | 1.1 ± 0.1* | 1.9 ± 0.1** | 0.9 ± 0.1†† | 1.1 ± 0.1 | 1.0 ± 0.1 | 2.6 ± 0.3** | 1.2 ± 0.2†† |
| HPRT1 | 1.1 ± 0.1 | 1.1 ± 0.1 | 0.9 ± 0.2 | 1.2 ± 0.1* | 1.1 ± 0.1† | 1.6 ± 0.1** | 1.3 ± 0.2* |
| DHODH | 1.0 ± 0.2 | 0.8 ± 0.1 | 0.9 ± 0.2† | 1.1 ± 0.1 | 1.0 ± 0.1 | 1.3 ± 0.1** | 1.2 ± 0.1 |
| UMPS | 1.2 ± 0.1* | 1.6 ± 0.1* | 1.0 ± 0.2†† | 1.1 ± 0.1 | 0.8 ± 0.2† | 2.4 ± 0.1* | 1.3 ± 0.1*††† |
| RRM1 | 1.0 ± 0.1* | 1.6 ± 0.3* | 0.8 ± 0.2† | 1.1 ± 0.1 | 1.0 ± 0.1† | 2.2 ± 0.2* | 1.1 ± 0.1*††† |
| TP | 1.5 ± 0.1* | 2.5 ± 0.1*** | 1.7 ± 0.4*† | 4.8 ± 2.2* | 1.8 ± 0.1*††† | 11.0 ± 3.4* | 2.5 ± 0.1***† |
| UPP1 | 1.1 ± 0.1* | 1.4 ± 0.4 | 0.7 ± 0.3† | 1.4 ± 0.1* | 0.8 ± 0.2† | 2.4 ± 0.9* | 1.4 ± 0.2† |
| UCK2 | 1.2 ± 0.1 | 1.3 ± 0.1* | 1.0 ± 0.2 | 1.2 ± 0.1 | 1.1 ± 0.1 | 1.9 ± 0.1** | 1.4 ± 0.2 |
| TK1 | 0.9 ± 0.3* | 1.6 ± 0.1* | 0.9 ± 0.2† | 1.2 ± 0.1 | 1.1 ± 0.1† | 2.3 ± 0.1** | 1.5 ± 0.2*† |
| DCK | 1.0 ± 0.1 | 1.6 ± 0.1* | 0.6 ± 0.2† | 0.9 ± 0.2 | 1.0 ± 0.1 | 2.6 ± 0.7* | 1.2 ± 0.3† |
| Oxidative stress | |||||||
| GPX1 | 2.7 ± 2.4 | 5.2 ± 3.5 | 3.6 ± 1.7 | 4.6 ± 2.7 | 4.4 ± 3.1 | 8.3 ± 6.1 | 5.0 ± 3.8 |
| SOD1 | 1.1 ± 0.1 | 2.1 ± 0.1** | 0.9 ± 0.1††† | 1.4 ± 0.1* | 0.8 ± 0.3† | 2.1 ± 0.5* | 1.1 ± 0.4* |
| SOD2 | 0.9 ± 0.4 | 2.1 ± 0.3* | 1.4 ± 0.2† | 1.1 ± 0.2 | 1.8 ± 0.2*† | 2.6 ± 0.1*** | 1.9 ± 1.1* |
| DNA repair | |||||||
| UNG1 | 1.1 ± 0.1* | 0.9 ± 0.1 | 1.0 ± 0.2 | 1.0 ± 0.1 | 1.1 ± 0.1 | 2.0 ± 0.2* | 1.3 ± 0.2*† |
| OGG1 | 1.2 ± 0.1* | 1.6 ± 0.2* | 1.0 ± 0.2† | 1.3 ± 0.1* | 1.3 ± 0.1* | 3.1 ± 0.3** | 1.8 ± 0.2**† |
| APE1 | 1.1 ± 0.1 | 1.8 ± 0.1* | 0.9 ± 0.2†† | 1.2 ± 0.1* | 1.1 ± 0.1 | 2.4 ± 0.2** | 1.3 ± 0.2†† |
Values represent the fold difference (±SD) of mRNA copies after 10 days of HepG2 treatment with agents by comparison with unexposed cells. Three independent cell cultures were measured per agent added. Significance levels:
P < 0.05,
P < 0.01,
P < 0.001 in comparison to controls (no uridine and no stavudine),
P < 0.05,
P < 0.01,
P < 0.001 in comparison to treatment without uridine. Bold entries represent significant up-regulation by a minimum factor of 2.
Gene abbreviations: LMNB1, lamin B1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ACTB, actin beta; 36B4, ribosomal phosphoprotein P0; PCNA, proliferating cell nuclear antigen; UbcH10, ubiquitin-conjugating enzyme H10; p21, cyclin-dependent kinase inhibitor 1A; CDKN2A, cyclin-dependent kinase inhibitor 2A; BAD, BCL2 antagonist of cell death; BCL2, B-cell lymphoma protein 2; BCL2L1, BCL antagonist of cell death; BAX, BCL2-associated X protein; FAS, fast apoptotic stimulus; FASLG, Fas ligand; CASP3, caspase 3; CASP8, caspase 8; CASP9, caspase 9; COX IV, cytochrome c oxidase subunit IV; ATP9, ATP synthase subunit 9; 16S rRNA; ND1, NADH dehydrogenase subunit 1, complex I; COX I, cytochrome c-oxidase subunit 1; ATP6, ATP synthase subunit 6; ND6, NADH dehydrogenase subunit 6; POLG, polymerase-γ (catalytic subunit); PGC-1, peroxisome proliferator-activated γ co-activator-1; TFAM, mitochondrial transcription factor A; DHFR, dihydrofolate reductase; DGUOK, deoxyguanosine kinase; HPRT1, hypoxanthine phosphoribosyltransferase 1; DHODH, dihydroorotate dehydrogenase; UMPS, uridine monophosphate synthetase; RRM1, ribonucleotide reductase M1; TP, thymidine phosphorylase; UPP1, uridine phosphorylase 1; UCK2, uridine-cytidine kinase 2; TK1, thymidine kinase 1; DCK, deoxycytidine kinase; GPX1, glutathione peroxidase 1; SOD1, superoxide dismutase 1; SOD2, superoxide dismutase 2; UNG1, uracil-DNA glycosylase; OGG1, 8-oxoguanine DNA glycosylase; APE1, apurinic/apyrimidinic endonuclease 1.
Statistics
All cell cultures were performed in triplicates. Each of these three completely independent cultures was used for each biomarker measurement at each time point. Group means were compared by analysis of variance, followed by unpaired t-tests. Graphics and statistical calculations were performed using the Sigma Plot 2000, version 8.0 (SPSS Inc., Chicago, IL) and the Sigma Stat, version 3.1 (Jandel Inc., San Rafael, CA) software packages.
Results
Hepatocyte Proliferation
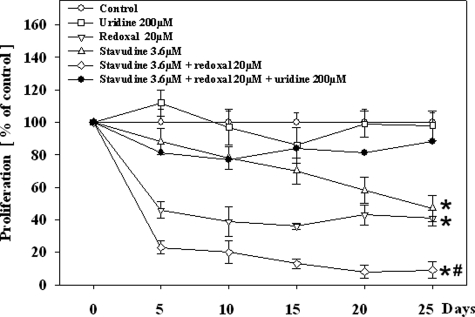
The HepG2 cells proliferated rapidly in medium without any additional agent. At the time of each harvest, the 2.7 × 106 cells seeded had doubled approximately every 2 days, resulting in 1.5 × 107 cells (SD, 3.0 × 106). Redoxal (20 μmol/L) approximately halved the rate of cell proliferation at each harvest (Figure 2). Stavudine (3.6 μmol/L) in contrast led to a time-dependent decrease of cell proliferation. The addition of redoxal to stavudine significantly enhanced the time-dependent inhibition of cell division by stavudine, whereas uridine (200 μmol/L) restored cell proliferation under co-treatment with redoxal alone, stavudine alone, and the combination of the two agents. Phase contrast microscopy performed after 25 days of exposure demonstrated abundant viable cells with all exposures. Cells treated with stavudine and/or redoxal without uridine appeared to be less abundant and slightly larger compared to their uridine-co-treated counterparts. Cell size however was not formally quantified.
Figure 2.
Cell proliferation in HepG2 cells under treatment with different agents. Each value represents the mean ± SE of three independent cultures. *P < 0.001 versus control; #P < 0.01 versus stavudine without uridine.
mtDNA, mtDNA-Encoded Respiratory Chain Subunits, and Intracellular ATP
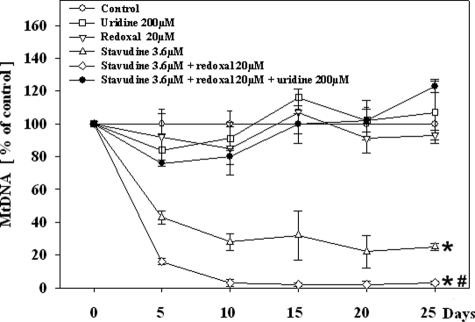
HepG2 cells exposed to stavudine developed a time-dependent mtDNA depletion, temporally preceding the decrease of cell proliferation (Figure 3). Whereas redoxal alone did not alter mtDNA levels, it accelerated the decline of mtDNA copy numbers in cells co-treated with stavudine. Uridine fully abrogated the onset of mtDNA depletion induced by stavudine, and by the combination of redoxal plus stavudine.
Figure 3.
mtDNA copies in HepG2 cells under treatment with different agents. Each value represents the mean measurement ± SE of three independent cultures. *P < 0.001 versus control; #P < 0.01 versus stavudine without uridine.
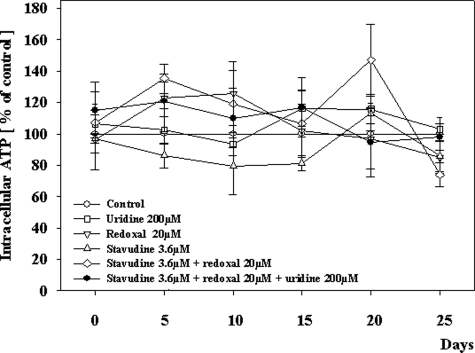
The expression of the mtDNA-encoded respiratory chain subunit COXII (normalized for COXIV) was not altered with redoxal (98% of control values; SD, 16%) and only slightly altered after the first 10 days of incubation with stavudine (81%; SD, 18%), or redoxal plus stavudine (80%; SD, 17%). After day 15 however and thus in coincidence with the reduction of cell counts but after the onset of mtDNA depletion, the expression of COXII was reduced in cells exposed to stavudine, and stavudine plus redoxal (40% of control values; SD, 23%; P = 0.075; and 37% of control values; SD, 21%; P = 0.07. respectively). Uridine prevented COXII depletion and fully normalized COXII expression in cells incubated with stavudine and stavudine plus redoxal. The application of redoxal alone did not alter COXII expression in mitochondria during the whole 25-day incubation period. In contrast to the profound mtDNA depletion, there was no significant difference between the treatments with respect to free ATP within the HepG2 cells at all time points (Figure 4).
Figure 4.
Intracellular ATP in HepG2 cells under treatment with different agents. Each value represents the mean measurement ± SE of three independent cultures. *P < 0.001 versus control; #P < 0.01 versus stavudine without uridine.
Lipid Content
The low concentration of stavudine that we used in the experiments described above did in not alter intracellular lipid content. The use of higher concentrations of stavudine (36 μmol/L) however, or 25-day exposure, strongly induced intracellular steatosis (not shown), as previously published.12 Redoxal also led to a time-dependent enhancement of intracellular lipids that was further enhanced in the combination of redoxal with stavudine (not shown). In all settings, lipid accumulation was fully prevented by the addition of uridine.12
Lipid Peroxidation
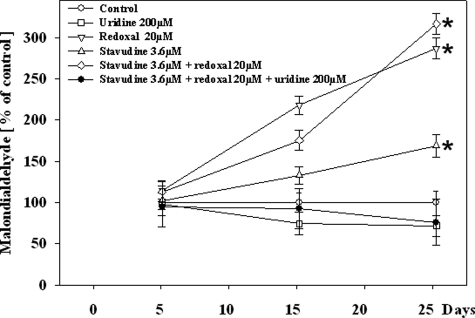
The generation of reactive oxygen species is an unavoidable byproduct of mitochondrial electron transport.29 Malondialdehyde production, an indicator of oxidative stress and production of free radicals was enhanced by stavudine and to a greater extent by redoxal. Uridine fully abrogated the effects of both agents on lipid peroxidation (Figure 5).
Figure 5.
Malondialdehyde content in HepG2 cells under treatment with different agents. Each value represents the mean measurement ± SE of three independent cultures. *P < 0.001 versus control; #P < 0.01 versus stavudine without uridine.
Caspase 3 Activation
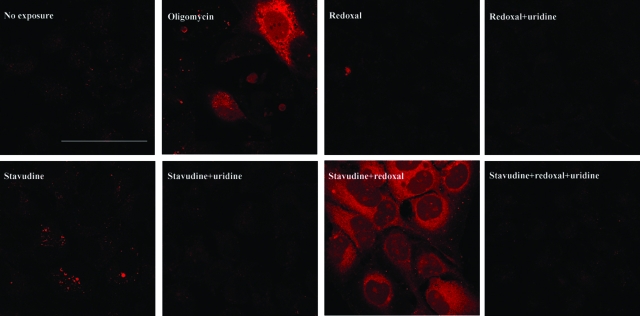
Animal models indicate enhanced hepatic apoptosis when liver mtDNA is depleted.6,30 We tested if putative indirect DHODH inhibition by stavudine may have effects on apoptosis similar to direct DHODH inhibition by redoxal. By analyzing the activation of the effector caspase 3, we found redoxal not to activate this protease (Figure 6). Stavudine also activated caspase 3 only minimally. Combining redoxal with stavudine however clearly increased the activation of caspase 3. Uridine fully prevented caspase 3 activation by stavudine and redoxal, alone and in combination.
Figure 6.
In vivo staining of activated caspase 3 in HepG2 cells with Red-DEVD-FMK after 25 days of cultivation. The redoxal concentration used was 20 μmol/L; the stavudine and uridine concentrations were 3.4 μmol/L and 200 μmol/L, respectively. Twenty μg/ml of oligomycin for 24 hours was used as a positive control for apoptosis. Pictures were taken at identical magnifications. Scale bar = 80 μm.
Nuclear and Mitochondrial Transcripts
To characterize the impact of redoxal and stavudine on cell cycle, we analyzed several transcripts involved in cell cycle checkpoint regulation (Table 2). p21 is a protein that promotes cell cycle arrest by binding to and inhibiting cyclin-dependent kinases (cdk), which normally, when coupled with specific cyclins, facilitate the orderly procession of the cell cycle. p21 accumulation thus impairs the activity of cyclin-dependent kinases, and promotes cell cycle arrest in G1.31 CDKN2A induces cell cycle arrest at G1 by preventing exit from G1 phase.32 We found that redoxal and stavudine induced an up-regulation of p21 and CDKN2A transcription. Other genes involved in cell cycling (proliferating cell nuclear antigen, UbcH10) were not changed by both substances. The combination of stavudine with redoxal led to a synergistic up-regulation of p21 and CDKN2A, two genes implicated in an arrest at the G1 phase of the cell cycle.32,33 This effect was attenuated by uridine.
The induction of apoptosis is governed by an elaborate array of checks and balances in which mitochondria participate.25,34 Intrinsic and extrinsic pathways to caspase-dependent cell death have been identified. Extrinsic apoptosis typically involves extracellular death receptors such as FAS, and recruitment of caspase 8. In intrinsic apoptosis, cellular stress is sensed by Bad and other proteins that then initiate Bax-dependent mitochondrial outer membrane permeabilization, resulting in caspase 9 and caspase 3 activation. Opening of the mitochondrial channel is antagonized by members of the Bcl-2 family. Redoxal and stavudine affected the transcription of many key players of apoptosis. Both agents participated synergistically in inducing caspase 3 transcription. Redoxal increased transcripts involved in the extrinsic (FAS, FASLG, and caspase 8) and intrinsic (Bad, caspase 3) pathway of apoptosis. The combination of redoxal with stavudine led to a further, mostly additive increase of gene transcripts compared to each treatment alone. Uridine normalized or attenuated these effects, but when given alone, had no action on gene transcripts involved in apoptosis.
Among the genes involved in mitochondrial energy metabolism, we observed a 4.1-fold increase in 16S rRNA transcripts with stavudine at day 10. Despite mtDNA depletion, the combination of redoxal with stavudine was found to synergistically up-regulate mtDNA encoded respiratory chain transcripts (ND1, COXI, ATP6, and ND6). nDNA encoded mtDNA transcripts (COXIV, ATP9) were also up-regulated. Similarly, polymerase-γ transcripts and the peroxisome-proliferator-activated receptor-γ co-activator (PGC1), a molecule involved in mitochondrial biogenesis and gluconeogenesis were found to be induced. Notably, the mitochondrial transcription factor A (TFAM) that binds to mtDNA as a chaperone35 was not up-regulated. Uridine fully normalized the transcript profile of all genes involved in energy metabolism.
Many enzymes participate in the pyrimidine and purine de novo synthesis, interconversion and salvage (Figure 1). The application of stavudine or redoxal most notably increased the steady state levels of thymidine phosphorylase (TP) transcripts. TP participates in the salvage of thymine, but is also responsible for the catabolism of thymidine analogues. The other gene transcripts tested were not affected. When we combined redoxal with stavudine, most genes related to nucleotide metabolism were up-regulated in a more than additive manner, indicating synergistic toxicity. Among the enzymes participating in the de novo synthesis of pyrimidines, UMPS and DHFR, but not DHODH were up-regulated. Genes responsible for pyrimidine salvage (UPP1, UCK2, TP, TK1, DCK) were also induced, as was DGUOK, an enzyme that regulates the intramitochondrial supply of purines and which is implicated in a hepatocerebral mtDNA depletion syndrome.36 The addition of uridine to the combination of stavudine plus redoxal reverted transcript levels.
We also analyzed the transcription of three genes participating in the scavenging of reactive oxygen species (GPX1, SOD1, SOD2). At the transcriptional level, stavudine had no significant effects at day 10, whereas redoxal up-regulated cytosolic and mitochondrial superoxide dismutase (SOD1, SOD2). The combination of redoxal with stavudine, slightly enhanced SOD2 expression. Uridine normalized transcripts associated with radical scavenging.
We finally studied the synergistic action of redoxal and stavudine with respect to transcripts involved in DNA repair. Oxidative base lesions of purines and pyrimidines in nuclear DNA and mtDNA are mainly repaired by base excision.37 The major enzyme removing oxidized pyrimidine in mitochondria DNA is UNG1.38,39 Oxidized purines are removed by OGG1.37 After the release of damaged bases, the resulting apurinic/apyrimidinic sites are cleaved by APE1 in the nucleus as well as in mitochondria.38 Redoxal induced a slight up-regulation of genes related to base excision repair, whereas stavudine had no impact. Combining redoxal with stavudine specifically increased mitochondrial pyrimidine (UNG1) and purine (OGG1) repair, as well as APE1. Although uridine alone had no effects, it virtually normalized the expression of repair transcripts when given with both other agents.
Discussion
We examined the hepatotoxicity of stavudine, a pyrimidine analogue reverse transcriptase and polymerase-γ inhibitor and focused on the role of diminished intracellular pyrimidine pools as a possible consequence of, and sensitizer for, mtDNA depletion. At the same time, we also investigated the effects of uridine in abrogating this mitochondrial cytotoxicity.
With the use of redoxal, an inhibitor of de novo pyrimidine synthesis, we demonstrated that pyrimidine removal sensitizes HepG2 cells toward stavudine-induced mtDNA depletion, an effect that preceded, or was paralleled by multiple other cellular events. The fact that the toxicological profile of stavudine and redoxal was similar and was either attenuated or almost completely antagonized by uridine as one single agent strongly suggests that pyrimidine depletion plays an important intermediary role and possibly represents even a common denominator in this form of toxicity. Our findings thus support a vicious circle according to which stavudine-mediated mtDNA and pyrimidine depletion allows for an accelerated mtDNA depletion by the same agent. The importance of pyrimidine depletion for the adverse cellular effects of stavudine is also underlined by the fact that the gene expression was multiplicative for the genes involved in nucleotide metabolism, whereas it was mostly additive for other pathways. Interestingly, some genes involved in purine nucleotide metabolism (DGUOK) were also up-regulated, possibly reflecting the fact that inhibition of the respiratory chain also causes a breakdown in purine triphosphates.18
We also investigated the effects of pyrimidine replenishment by uridine on other cellular pathways. From studies with leflunomide, another direct DHODH inhibitor, there is evidence that low intracellular pyrimidine pools trigger and mediate apoptosis.33,40 In our experiments we were not able to detect apoptosis in terms of caspase 3 activation with either stavudine or redoxal alone because we specifically selected the concentration of each agent to only diminish cell proliferation by 50%. In the combination of redoxal and stavudine however, we detected a clear activation of the effector caspase 3 and an up-regulation of multiple gene transcripts involved in apoptosis. Similar proapoptotic effects including caspase 3 activation by hepatic mtDNA depletion have also been observed in vivo.6,30 In light of the preserved ATP levels and the antagonism by uridine, our results suggest that pyrimidine depletion and not the lack of chemical energy is the intracellular harbinger of apoptosis. In addition our results suggest that the pyrimidine depletion by stavudine results in a cell cycle arrest at G1, as indicated by the up-regulation of p21 and CDKN2A. Such G1 arrest was previously suggested to also result from direct DHODH inhibition.33,41
With stavudine exposure, the mtDNA-encoded respiratory chain transcripts did not change despite a 50% reduction in mtDNA copy numbers at the same time point. Thus, the preservation of respiratory chain protein (COXII) results at least in part from compensation at the transcriptional level. The simultaneous up-regulation of 16S rRNA may be interpreted as an additional translational effort for compensation. Both mechanisms may act in concert and explain in part the sigmoidal genotype phenotype correlation, commonly referred to as threshold effect.42
Redoxal alone also enhanced some nuclear transcripts involved in energy metabolism and mitochondrial biogenesis. To the same end, leflunomide was previously found to induce the proliferation of mitochondria.43 This is best explained by the cross talk between mitochondria and nucleus. For example, mice deficient in ANT1 (a mitochondrial nucleoside transporter) compensate for the lack of cytosolic ATP by increasing mitochondrial transcripts related to oxidative phosphorylation.44 How redoxal enhances oxidative stress is unknown. The fact that malondialdehyde levels of cells treated with redoxal alone were similar to those treated with redoxal plus stavudine may indicate that oxidative stress does not account for the observed induction of apoptosis under the combination of redoxal with stavudine.
Our results have several clinically important implications. In HIV patients treated with stavudine, therapeutic agents that have an impact on pyrimidine pools should be avoided. Such substances are leflunomide, but may also be inhibitors of tetrahydrofolate reductase such as methotrexate, aminopterin, pyrimethamine, and trimethoprim. A metabolite of the anti-herpes drug brivudin inhibits dihydropyrimidine dehydrogenase and thus may adversely affect the intracellular balance of natural pyrimidines and stavudine.45 Gender-specific differences in pyrimidine pools may contribute to the higher incidence of lactic acidosis in females.11 An interaction with the intramitochondrial availability of pyrimidines may also explain the observation that some NRTIs increase the mitochondrial toxicity of other NRTIs, although they by themselves have no impact on polymerase-γ.2,46 Zidovudine for example does not induce hepatic mtDNA depletion in vitro and in vivo5,47 but instead may limit pyrimidine supply by being an inhibitor of thymidine kinases.48 In accord with the importance of the availability of intracellular pyrimidines, the toxicity of zidovudine can be antagonized by uridine.12,13,17,49
In summary, our findings suggest that the intracellular depletion of pyrimidine metabolites sensitizes for mtDNA depletion and is also an important intermediary between mtDNA depletion and multiple secondary adverse effects. Pyrimidine depletion also provides an explanation of the mitochondrial toxicity of some agents, which do not inhibit mtDNA replication. Uridine supplementation has no intrinsic effects on cell metabolism, but is a promising strategy in the treatment of side effects related to pyrimidine depletion. Drugs, which have an impact on pyrimidine pools, should be avoided in HIV patients treated with stavudine.
Footnotes
Address reprint requests to Prof. Dr. Ulrich A. Walker, Medizinische Universitätsklinik, Dept. of Rheumatology and Clinical Immunology, Hugstetterstr. 55, D-79106 Freiburg, Germany. E-mail: ulrich.walker@klinikum. uni-freiburg.de.
Supported by the Deutsche Forschungsgemeinschaft (grant no. Wa1387/1-5).
References
- Brinkman K, Smeitink JA, Romijn JA, Reiss P. Mitochondrial toxicity induced by nucleoside-analogue reverse-transcriptase inhibitors is a key factor in the pathogenesis of antiretroviral-therapy-related lipodystrophy. Lancet. 1999;354:1112–1115. doi: 10.1016/S0140-6736(99)06102-4. [DOI] [PubMed] [Google Scholar]
- Walker UA, Setzer B, Venhoff N. Increased long-term mitochondrial toxicity in combinations of nucleoside analogue reverse-transcriptase inhibitors. AIDS. 2002;16:2165–2173. doi: 10.1097/00002030-200211080-00009. [DOI] [PubMed] [Google Scholar]
- Gerschenson M, Nguyen VT, St Claire MC, Harbaugh SW, Harbaugh JW, Proia LA, Poirier MC. Chronic stavudine exposure induces hepatic mitochondrial toxicity in adult Erythrocebus patas monkeys. J Hum Virol. 2001;4:335–342. [PubMed] [Google Scholar]
- Lewis W, Dalakas MC. Mitochondrial toxicity of antiviral drugs. Nat Med. 1995;1:417–422. doi: 10.1038/nm0595-417. [DOI] [PubMed] [Google Scholar]
- Walker UA, Bäuerle J, Laguno M, Murillas J, Mauss S, Schmutz G, Setzer B, Miquel R, Gatell JM, Mallolas J. Depletion of mitochondrial DNA in liver under antiretroviral therapy with didanosine, stavudine, or zalcitabine. Hepatology. 2004;39:311–317. doi: 10.1002/hep.20074. [DOI] [PubMed] [Google Scholar]
- Lebrecht D, Vargas Infante YA, Setzer B, Kirschner J, Walker UA. Uridine supplementation antagonizes zalcitabine-induced microvesicular steatohepatitis in mice. Hepatology. 2007;15:72–79. doi: 10.1002/hep.21490. [DOI] [PubMed] [Google Scholar]
- Lambert JS, Seidlin M, Reichman RC, Plank CS, Laverty M, Morse GD, Knupp C, McLaren C, Pettinelli C, Valentine FT. 2′,3′-dideoxyinosine (ddI) in patients with the acquired immunodeficiency syndrome or AIDS-related complex. A phase I trial. N Engl J Med. 1990;322:1333–1340. doi: 10.1056/NEJM199005103221901. [DOI] [PubMed] [Google Scholar]
- Morris AA, Taanman JW, Blake J, Cooper JM, Lake BD, Malone M, Love S, Clayton PT, Leonard JV, Schapira AH. Liver failure associated with mitochondrial DNA depletion. J Hepatol. 1998;28:556–563. doi: 10.1016/s0168-8278(98)80278-x. [DOI] [PubMed] [Google Scholar]
- Chariot P, Drogou I, de Lacroix-Szmania I, Eliezer-Vanerot MC, Chazaud B, Lombes A, Schaeffer A, Zafrani ES. Zidovudine-induced mitochondrial disorder with massive liver steatosis, myopathy, lactic acidosis, and mitochondrial DNA depletion. J Hepatol. 1999;30:156–160. doi: 10.1016/s0168-8278(99)80020-8. [DOI] [PubMed] [Google Scholar]
- Gerard Y, Maulin L, Yazdanpanah Y, Tribonniere X, Amiel C, Maurage CA, Robin S, Sablonniere B, Dhennain C, Mouton Y. Symptomatic hyperlactataemia: an emerging complication of antiretroviral therapy. AIDS. 2000;14:2723–2730. doi: 10.1097/00002030-200012010-00012. [DOI] [PubMed] [Google Scholar]
- Bolhaar MG, Karstaedt AS. A high incidence of lactic acidosis and symptomatic hyperlactatemia in women receiving highly active antiretroviral therapy in Soweto. South Africa Clin Infect Dis. 2007;45:254–260. doi: 10.1086/518976. [DOI] [PubMed] [Google Scholar]
- Walker UA, Venhoff N, Koch E, Olschweski M, Schneider J, Setzer B. Uridine abrogates mitochondrial toxicity related to nucleoside analogue reverse transcriptase inhibitors in HepG2 cells. Antivir Ther. 2003;8:463–470. [PubMed] [Google Scholar]
- Walker UA, Auclair M, Lebrecht D, Kornprobst M, Capeau J, Caron M. Uridine abrogates the adverse effects of antiretroviral pyrimidine analogues on adipose cell functions. Antivir Ther. 2006;11:25–34. [PubMed] [Google Scholar]
- Walker UA, Venhoff N. Uridine in the prevention and treatment of NRTI-related mitochondrial toxicity. Antivir Ther. 2005;10:M117–M123. [PubMed] [Google Scholar]
- Walker UA, Langmann P, Miehle N, Zilly M, Klinker H, Petschner F. Beneficial effects of oral uridine in mitochondrial toxicity. AIDS. 2004;18:1085–1086. doi: 10.1097/00002030-200404300-00025. [DOI] [PubMed] [Google Scholar]
- Banasch M, Goetze O, Knyhala K, Potthoff A, Schlottmann R, Kwiatek MA, Bulut K, Schmitz F, Schmidt WE, Brockmeyer NH. Uridine supplementation enhances hepatic mitochondrial function in thymidine-analogue treated HIV-infected patients. AIDS. 2006;20:1554–1556. doi: 10.1097/01.aids.0000237373.38939.14. [DOI] [PubMed] [Google Scholar]
- Sutinen J, Walker UA, Häkkinen AM, Ristola M, Yki-Jarvinen H. Uridine for the treatment of HAART-associated lipodystrophy—a randomized, double-blind, placebo-controlled trial. Antivir Ther. 2007;12:97–105. [PubMed] [Google Scholar]
- Gattermann N, Dadak M, Hofhaus G, Wulfert M, Berneburg M, Loeffler ML, Simmonds HA. Severe impairment of nucleotide synthesis through inhibition of mitochondrial respiration. Nucleosides Nucleotides Nucleic Acids. 2004;23:1275–1279. doi: 10.1081/NCN-200027545. [DOI] [PubMed] [Google Scholar]
- Kakuda TN. Pharmacology of nucleoside and nucleotide reverse transcriptase inhibitor-induced mitochondrial toxicity. Clin Ther. 2000;22:685–708. doi: 10.1016/S0149-2918(00)90004-3. [DOI] [PubMed] [Google Scholar]
- Knecht W, Löffler M. Redoxal as a new lead structure for dihydroorotate dehydrogenase inhibitors: a kinetic study of the inhibition mechanism. FEBS Lett. 2000;467:27–30. doi: 10.1016/s0014-5793(00)01117-0. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setzer B, Schlesier M, Walker UA. Effects of didanosine-related depletion of mtDNA in human T lymphocytes. J Infect Dis. 2005;191:848–855. doi: 10.1086/427655. [DOI] [PubMed] [Google Scholar]
- Frings CS, Fendley TW, Dunn RT, Queen CA. Improved determination of total serum lipids by the sulfo-phospho-vanillin reaction. Clin Chem. 1972;18:673–674. [PubMed] [Google Scholar]
- Tüzgen S, Kaynar MY, Guner A, Gumustas K, Belce A, Etus V, Ozyurt E. The effect of epidural cooling on lipid peroxidation after experimental spinal cord injury. Spinal Cord. 1998;36:654–657. doi: 10.1038/sj.sc.3100660. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Martin SJ. Caspase-independent cell death. Nat Med. 2005;11:725–730. doi: 10.1038/nm1263. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg L, Bjorck E, Hashemi J, Zedenius J, Hoog A, Farnebo LO, Reimers M, Larsson C. Distinction in gene expression profiles demonstrated in parathyroid adenomas by high-density oligoarray technology. Eur J Endocrinol. 2005;152:459–470. doi: 10.1530/eje.1.01864. [DOI] [PubMed] [Google Scholar]
- Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge JM, Halliwell B. Free radicals and antioxidants in the year 2000. A historical look to the future. Ann NY Acad Sci. 2000;899:136–147. doi: 10.1111/j.1749-6632.2000.tb06182.x. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Haouzi D, Descatoire V, Demeilliers C, Sutton A, Vadrot N, Fromenty B, Feldmann G, Pessayre D, Berson A. Tacrine inhibits topoisomerases and DNA synthesis to cause mitochondrial DNA depletion and apoptosis in mouse liver. Hepatology. 2003;38:715–725. doi: 10.1053/jhep.2003.50353. [DOI] [PubMed] [Google Scholar]
- Afshari CA, Nichols MA, Xiong Y, Mudryj M. A role for a p21–E2F interaction during senescence arrest of normal human fibroblasts. Cell Growth Differ. 1996;7:979–988. [PubMed] [Google Scholar]
- Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- Fox RI. Mechanism of action of leflunomide in rheumatoid arthritis. J Rheumatol. 1998;53:S20–S26. [PubMed] [Google Scholar]
- Kass GE. Mitochondrial involvement in drug-induced hepatic injury. Chem Biol Interact. 2006;163:145–159. doi: 10.1016/j.cbi.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Saada-Reisch A. Deoxyribonucleoside kinases in mitochondrial DNA depletion. Nucleosides Nucleotides Nucleic Acids. 2004;23:1205–1215. doi: 10.1081/NCN-200027480. [DOI] [PubMed] [Google Scholar]
- Slupphaug G, Kavli B, Krokan HE. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat Res. 2003;531:231–251. doi: 10.1016/j.mrfmmm.2003.06.002. [DOI] [PubMed] [Google Scholar]
- Akbari M, Otterlei M, Pena-Diaz J, Krokan HE. Different organization of base excision repair of uracil in DNA in nuclei and mitochondria and selective upregulation of mitochondrial uracil-DNA glycosylase after oxidative stress. Neuroscience. 2007;145:1201–1212. doi: 10.1016/j.neuroscience.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Otterlei M, Haug T, Nagelhus TA, Slupphaug G, Lindmo T, Krokan HE. Nuclear and mitochondrial splice forms of human uracil-DNA glycosylase contain a complex nuclear localisation signal and a strong classical mitochondrial localisation signal, respectively. Nucleic Acids Res. 1998;26:4611–4617. doi: 10.1093/nar/26.20.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren M. Regulation of the p53 tumor suppressor protein. J Biol Chem. 1999;274:36031–36034. doi: 10.1074/jbc.274.51.36031. [DOI] [PubMed] [Google Scholar]
- Rückemann K, Fairbanks LD, Carrey EA, Hawrylowicz CM, Richards DF, Kirschbaum B, Simmonds HA. Leflunomide inhibits pyrimidine de novo synthesis in mitogen-stimulated T-lymphocytes from healthy humans. J Biol Chem. 1998;273:21682–21691. doi: 10.1074/jbc.273.34.21682. [DOI] [PubMed] [Google Scholar]
- Clark KM, Taylor RW, Johnson MA, Chinnery PF, Chrzanowska-Lightowlers ZM, Andrews RM, Nelson IP, Wood NW, Lamont PJ, Hanna MG, Lightowlers RN, Turnbull DM. An mtDNA mutation in the initiation codon of the cytochrome C oxidase subunit II gene results in lower levels of the protein and a mitochondrial encephalomyopathy. Am J Hum Genet. 1999;64:1330–1339. doi: 10.1086/302361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spodnik JH, Wozniak M, Budzko D, Teranishi MA, Karbowski M, Nishizawa Y, Usukura J, Wakabayashi T. Mechanism of leflunomide-induced proliferation of mitochondria in mammalian cells. Mitochondrion. 2002;2:163–179. doi: 10.1016/s1567-7249(02)00045-4. [DOI] [PubMed] [Google Scholar]
- Murdock DG, Boone BE, Esposito LA, Wallace DC. Up-regulation of nuclear and mitochondrial genes in the skeletal muscle of mice lacking the heart/muscle isoform of the adenine nucleotide translocator. J Biol Chem. 1999;274:14429–14433. doi: 10.1074/jbc.274.20.14429. [DOI] [PubMed] [Google Scholar]
- De Clercq E. Discovery and development of BVDU (brivudin) as a therapeutic for the treatment of herpes zoster. Biochem Pharmacol. 2004;68:2301–2315. doi: 10.1016/j.bcp.2004.07.039. [DOI] [PubMed] [Google Scholar]
- Olivero OA, Fernandez JJ, Antiochos BB, Wagner JL, St Claire ME, Poirier MC. Transplacental genotoxicity of combined antiretroviral nucleoside analogue therapy in Erythrocebus patas monkeys. J Acquir Immune Defic Syndr. 2002;29:323–329. doi: 10.1097/00126334-200204010-00001. [DOI] [PubMed] [Google Scholar]
- Walker UA, Setzer B, Venhoff N. Increased long-term mitochondrial toxicity in pyrimidine nucleoside combinations. Antiviral Ther. 2001;6:13–14. [Google Scholar]
- Lynx MD, McKee EE. 3′-Azido-3′-deoxythymidine (AZT) is a competitive inhibitor of thymidine phosphorylation in isolated rat heart and liver mitochondria. Biochem Pharmacol. 2006;72:239–243. doi: 10.1016/j.bcp.2006.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommadossi JP, Carlisle R, Schinazi RF, Zhou Z. Uridine reverses the toxicity of 3′-azido-3′-deoxythymidine in normal human granulocyte-macrophage progenitor cells in vitro without impairment of antiretroviral activity. Antimicrob Agents Chemother. 1988;32:997–1001. doi: 10.1128/aac.32.7.997. [DOI] [PMC free article] [PubMed] [Google Scholar]