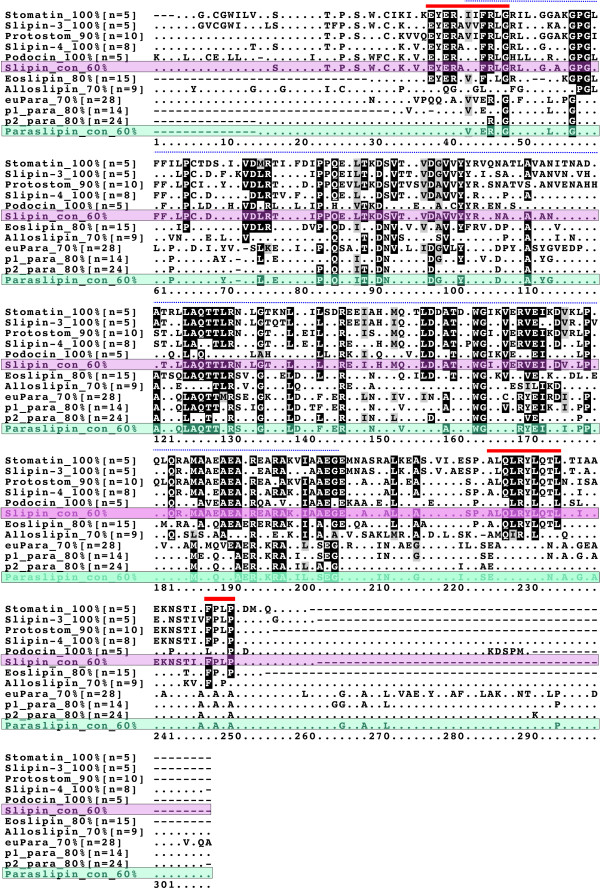
Figure 6.
Alignment of stomatin subfamily consensus sequences. Consensus sequences (100-70 %) were generated by selecting and aligning (ClustalX) subfamily members identified in monophyletic groups in Figures 1-4. The shading threshold was set to 0.6. Identical residues have a black background and similar residues have a grey background. A 60 % slipin (pink) and paraslipin (green) consensus sequence is also displayed. p2_para refers to prokaryotic paraslipins present in the upper clade in Figure 3, whilst p1_para refers to prokaryotic paraslipins present in the lower clade with eukaryotic paraslipins (euPara). The dotted line above the sequences shows the region shared by all stomatin family members, whilst the solid red line indicates regions shared by slipins and eoslipins to the exclusion of alloslipins. n = the number of sequences used to generate each consensus. – indicates the position of a gap, indicates an unconserved amino acid.