Abstract
Several Legionella pneumophila proteins were highly expressed in low-temperature supernatants. One of these proteins was the peptidyl-prolyl isomerase PpiB. Mutants lacking ppiB exhibited reduced growth at 17°C. Since PpiB lacked a signal sequence and was present in 17°C supernatants of type II and type IV secretion mutants, this protein may be secreted by a novel mechanism.
Legionella pneumophila is the agent of Legionnaires' disease pneumonia (14), and infection results from inhalation of contaminated water droplets from aerosol-generating devices (28). L. pneumophila occurs naturally in freshwaters (3, 15, 23, 29, 40), but it is also widespread in man-made water systems (26, 33, 45). It exists in the planktonic phase, in biofilms, and as an intracellular parasite of protozoans (12, 24, 25, 29, 31). But the distribution of L. pneumophila is also likely due to its capacity to survive at 4 to 63°C (15, 23, 44, 45). Thus, there is need for understanding L. pneumophila survival at low temperatures. Recently, we observed that L. pneumophila lsp mutants deficient in type II secretion grow normally at 30 to 37°C but their growth at 12 to 25°C is impaired (6, 22, 41). The wild type stimulates the growth of an lsp mutant at 25°C when they are plated near each other (41), suggesting that secreted factors promote low-temperature growth.
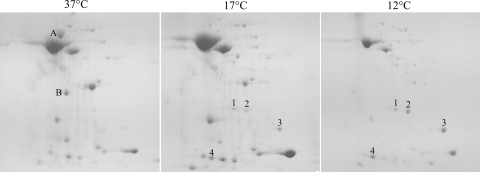
To identify secreted proteins that are newly expressed or hyperexpressed when the wild type is grown at low temperatures, strain 130b (= ATCC BAA-74) was grown to late log phase in buffered yeast extract (BYE) broth at 37, 17, and 12°C, and then filter-sterilized culture supernatants were examined by two-dimensional polyacrylamide gel electrophoresis, as previously described (10). Although growth slowed with decreasing temperature, there were many similarities between the profiles (Fig. 1), indicating that L. pneumophila secretes proteins while it is growing at 12 to 37°C. Low-temperature supernatants had slightly fewer proteins rather than more proteins, indicating that low-temperature incubation does not result in wholesale lysis. In addition to the similarities, there were differences. For example, the amounts of some proteins present at 37°C were reduced at 17 to 12°C (Fig. 1). There were also proteins that were more pronounced in the 17 and 12°C samples (e.g., spots 1 to 4 in Fig. 1). Thus, L. pneumophila secretion changes when the organisms is grown at low temperatures. We hypothesized that proteins whose amounts are greater at 17 to 12°C contribute to survival at low temperatures.
FIG. 1.
Two-dimensional polyacrylamide gel electrophoresis analysis of culture supernatants obtained from L. pneumophila grown at low temperatures. Wild-type strain 130b was grown in BYE broth to late log phase at 37, 17, or 12°C, and then the proteins in the different cell-free culture supernatants were separated by two-dimensional polyacrylamide gel electrophoresis and stained with Coomassie blue. Some protein spots whose amounts were greater in the 37°C sample are indicated by letters in the left panel, whereas some of the proteins that were more prominently expressed in the low-temperature samples are indicated by numbers in the center and right panels. Similar protein profiles were obtained on at least two other occasions using material derived from independent cultures.
To identify such proteins, spots 1 and 2 were analyzed by mass spectrometry (10), and the data were compared to a database (http://genolist.pasteur.fr/LegioList/). The 164-amino-acid protein in spot 1 was identified as PpiB (Lcy), a peptidyl-prolyl isomerase (PPIase) belonging to the cyclophilin family. ppiB is either monocistronic or the last gene in a small operon, and the preceding open reading frame encodes a tRNA ribosyltransferase. The 188-amino-acid protein in spot 2 was also annotated as a cyclophilin PPIase, and in the sequenced strains Philadelphia, Paris, and Lens the monocistronic genes that encode it are lpg1962, lpp1946, and lpl1936, respectively. Previously, PpiB was purified from cytoplasmic extracts of L. pneumophila Philadelphia-1 and demonstrated to have PPIase activity (37). In that study, however, there was no attempt to look for protein in supernatants or in bacteria grown at temperatures other than 37°C. There have been no previous studies of the putative PPIase of spot 2; thus, we refer to this protein as Lpg1962. Because PpiB and Lpg1962 were in wild-type supernatants that did not show evidence of cell lysis or leakage, these two proteins likely are secreted proteins. The fact that a previous study had shown that PpiB was present in cell extracts does not invalidate our findings. For example, cell-associated PpiB might simply represent a protein on its way toward secretion, or perhaps PpiB is maintained within and outside the cell. Alternatively, this protein might exist mainly in cells at 37°C but be secreted at 17°C. Lpg1962 has a signal sequence (16, 27) and thus is likely a substrate for type II secretion. PpiB lacks a typical signal sequence or a signal sequence with twin arginines (34), suggesting that its presence in supernatants depends on another mechanism.
To determine if PpiB and Lpg1962 promote growth at low temperatures, we used allelic exchange to construct mutants. First, primers were designed to amplify genes from 130b DNA; Mas23 (5′-CGTACGGAGCTCATATTCAG) and Mas25 (5′-TGGTAATATTTTCAATGACTACAGG) yielded a 773-bp fragment for lpg1962, and Mas26 (5′-TCTGCAATGAATACGGATGG) and Mas27 (5′-GGTACA CAAAAAGTTCTCGC) yielded a 1,552-bp fragment containing ppiB. Fragments containing ppiB and lpg1962 were ligated into pGEM-T Easy, yielding pB24 and pR11. pB24 was digested with BamHI, which cut 271 bp after the ppiB start codon, and was then ligated to a Kmr gene from pMB2190 (17) to obtain pB24K or to a Gmr gene from pX1918GT (39) to produce pB24G2. Next, NotI fragments of pB24K and pB24G2 containing the disrupted genes were cloned into the SmaI site of pRE112 (13) to obtain pB24KS3 and pB24GS4. pR11 was digested with AgeI, which cut 292 bp after the lpg1962 start, and then was ligated to the Kmr and Gmr cassettes, resulting in pR11K1 and pR11G2. Following NotI digestion, the disrupted lpg1962 genes were cloned into pRE112, yielding Kmr pR11K1S3 and Gmr pR11G2S3. 130b was transformed with pR11G2S3, pR11K1S3, pB24GS4, and pB24KS3 by electroporation (7), and mutants were selected as previously described (34). To construct lpg1962 ppiB double mutants, a Gmr lpg1962 mutant was transformed with pB24KS3, and a Kmr lpg1962 mutant was transformed with pB24GS4. Ultimately, six mutants were obtained: Gmr NU340 and Kmr NU341 for ppiB, Gmr NU342 and Kmr NU343 for lpg1962, and Kmr Gmr NU344 and NU345 for ppiB lpg1962. All mutants grew normally on buffered charcoal-yeast extract (BCYE) agar and in BYE broth at 37°C (Fig. 2A; data not shown), indicating that ppiB and lpg1962 are not required for extracellular growth under standard conditions. The fact that a ppiB mutant grows normally at 37°C was previously observed (37).
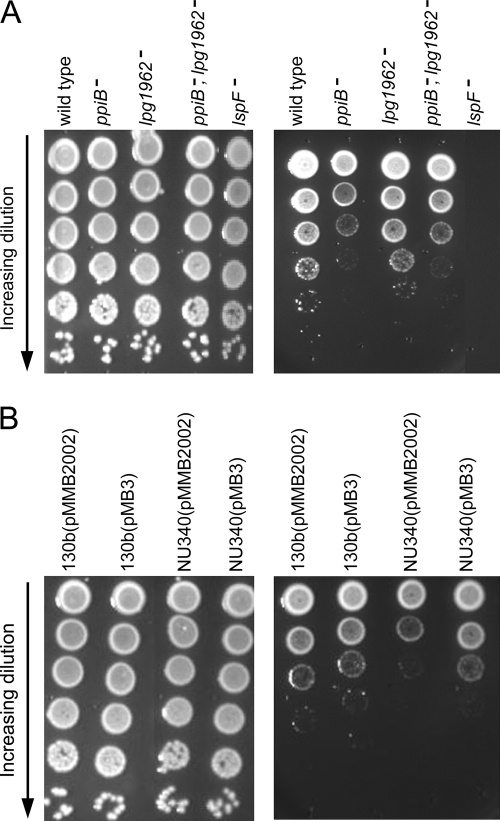
FIG. 2.
Low-temperature growth of the wild type and ppiB and lpg1962 mutants of L. pneumophila on BCYE agar. (A) Tenfold serial dilutions of wild-type strain 130b, ppiB mutant NU340, lpg1962 mutant NU342, ppiB lpg1962 double mutant NU344, and lspF mutant NU275 containing equivalent numbers of CFU (as determined at 37°C) were spotted onto BCYE agar plates and then incubated at either 37°C (left panel) or 17°C (right panel). Images of bacterial growth were then obtained after 3 days for the 37°C plates and after 20 days for the 17°C plates. The results are representative of at least three independent experiments. (B) Tenfold serial dilutions of wild-type strain 130b and ppiB mutant NU340 containing either the vector pMMB2002 or the complementing plasmid pMB3 (i.e., ppiB cloned into pMMB2002) were spotted onto BCYE agar and then incubated at 37°C (left panel) or 17°C (right panel). Images of bacterial growth were then obtained as described above. For unknown reasons, the vector alone slowed the growth of both the wild type and the mutant at 17°C (compared to the 17°C cultures used for panel A). Nonetheless, the specific growth-stimulating effect of cloned ppiB on NU340 at 17°C is still evident. The results are representative of two independent experiments.
Next, we compared the growth of 130b and the growth of the mutants on BCYE agar at 17°C (Fig. 2A). As hypothesized, ppiB mutant NU340 displayed reduced survival at this low temperature. Since independent ppiB mutant NU341 also showed impaired growth (data not shown), these data indicated that the defect was due to the loss of ppiB rather than second-site mutations. When an intact ppiB gene was reintroduced into NU340 on pMB3, the wild type and the ppiB mutant grew comparably (Fig. 2B). To create pMB3, ppiB was amplified using Mas27 and Mas28 (5′-TGTTTTGCATGATGTTTGTAAT) and cloned into pMMB2002 (35). In contrast to the results for ppiB, lpg1962 mutants NU342 and NU343 grew like the wild type when they were plated at low temperature (Fig. 2A; data not shown). Compatible with these results, the ppiB lpg1962 double mutants had reduced abilities to grow in the same way as the ppiB mutants (Fig. 2A; data not shown). To confirm the results obtained by plating, we compared the growth of 130b and the growth of mutants in BYE broth at 17°C. The ppiB mutants and the double mutants exhibited reduced growth, whereas lpg1962 mutants grew normally (Fig. 3A). After reintroduction of ppiB, the wild type and the ppiB mutants grew comparably in broth at 17°C (Fig. 3B). These data indicate that PpiB, but not Lpg1962, is necessary for optimal extracellular growth at low temperatures. The ppiB mutants were not as impaired at low temperatures as lspF mutant NU275 (Fig. 2A and 3A) (35, 41), suggesting that additional secreted factors promote low-temperature growth. Indeed, a ppiB mutant was able to stimulate the growth of the lspF mutant at a low temperature when these mutants were plated near each other (data not shown).
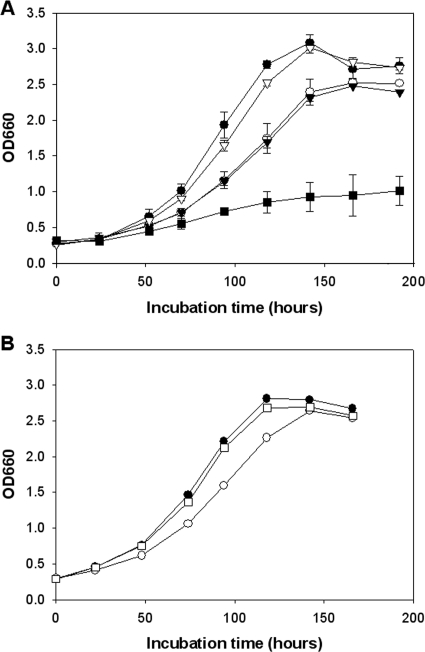
FIG. 3.
Low-temperature growth of the wild type and ppiB and lpg1962 mutants of L. pneumophila in BYE broth. Log-phase bacteria were inoculated into BYE broth at 17°C, and then the growth of the indicated strains was monitored by recording the optical densities of the cultures at various times. (A) Comparison of the growth of wild-type strain 130b (•), ppiB mutant NU340 (○), lpg1962 mutant NU342 (▿), ppiB lpg1962 double mutant NU344 (▾), and lspF mutant NU275 (▪). (B) Comparison of the growth of wild-type strain 130b (•), ppiB mutant NU340 (○), and complemented ppiB mutant NU340 (pMB3) (□). The apparent differences in growth between either the ppiB mutant or the ppiB lpg1962 mutant and the wild-type strain or the complemented ppiB mutant were statistically significant, as were the differences in growth between the lspF mutant and the other strains (P < 0.05, Student's t test). The slight differences in optical densities at 660 nm (OD660) between the wild type and the lpg1962 mutant observed at some time points were not seen in repeat experiments. The data are the means and standard deviations for duplicate cultures and are representative of at least three (A) and two (B) independent experiments.
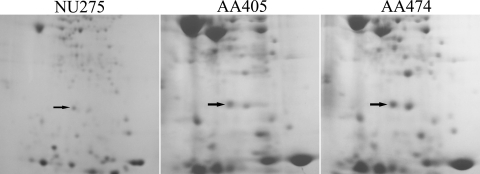
Because of the newfound importance of PpiB, we further investigated the mechanism by which this protein appears in supernatants. As noted previously, the absence of a Sec- or Tat-dependent signal sequence indicated that there is not type II secretion. Compatible with this, supernatants from an lspF mutant grown at 17°C contained PpiB (Fig. 4). But the presence of more proteins in the mutant supernatants than in wild-type supernatants (Fig. 1), rather than many fewer proteins, as observed when 37°C supernatants were compared (10), suggests that a type II mutant undergoes a greater degree of leakage or lysis at 17°C. L. pneumophila also possesses Lvh type IVA and Dot/Icm type IVB secretion (42). Proteins exported via type IV secretion often do not contain typical signal sequences (18). Thus, we examined supernatants obtained from dotG mutant AA405 and lvhB9 mutant AA474 (from Cary Engleberg, University of Michigan) grown at 17°C (Fig. 4). In both cases, PpiB was present, indicating that the PPIase does not require one of the type IV pathways for export. Neither type IV mutant exhibited reduced growth on BCYE agar or in BYE broth at 17°C (data not shown). These data suggest that PpiB is not released by one of the three known Legionella secretion systems. Genome sequencing has suggested the presence of a type I secretion system in L. pneumophila, as well as the presence of type V secretion in some strains (4, 21). However, PpiB lacks the glycine-rich repeats often present in type I substrates (11), and the “autotransporters” of type V secretion generally contain Sec-dependent signal sequences (18). L. pneumophila lacks type III secretion but does express flagella, which might provide a pathway for export (8, 20).
FIG. 4.
Two-dimensional polyacrylamide gel electrophoresis analysis of culture supernatants obtained from L. pneumophila type II and type IV secretion mutants grown at low temperatures. lspF mutant NU275, dotG mutant AA405, and lvhB9 mutant AA474 were grown in BYE broth to late log phase at 17°C, and then the proteins in the cell-free culture supernatants were separated by two-dimensional polyacrylamide gel electrophoresis and stained with Coomassie blue. The PpiB spot is indicated by an arrow.
Regardless of variations in the localization of PpiB or its secretion mechanism, our analyses indicate that PpiB is required for optimal growth at low temperatures. Given its PPIase activity (37), PpiB may catalyze the isomerization of secreted proteins, such as type II substrates, that promote survival at low temperatures. Alternatively, PpiB might assist cold-adapted exoenzymes as a chaperone (5, 19, 36, 38). Finally, PpiB, whether as an isomerase or as a chaperone, might promote the functioning of a secretion apparatus. There are examples of PPIases that are secreted and surface expressed by microbes, such as HP0175 of Helicobacter pylori and Mip of L. pneumophila (1, 9, 30), and there are PPIases that have ben previously linked to low-temperature adaptation, such as cell-associated PpiB of Bacillus subtilis, FKBP of Shewanella sp., and RotA of Erwinia chrysanthemi (2, 32, 43). But the connection that we uncovered between a secreted PPIase and low-temperature growth is a novel observation.
Previously, Schmidt et al. observed that a ppiB mutant has a reduced ability to grow in Acanthamoeba castellanii, an aquatic protozoan that serves as an intracellular niche for L. pneumophila (37). Thus, our data for the extracellular growth of ppiB mutants at low temperature indicate that PpiB likely promotes survival in natural habitats by at least two mechanisms. For Lpg1962, the absence of a growth defect in the lpg1962 mutant does not necessarily indicate irrelevance for low-temperature growth, since it is possible that another PPIase can replace Lpg1962. Combined, the present findings involving PpiB and previous work on Lsp indicate that a variety of secretion functions aid L. pneumophila, and perhaps other bacteria, in growing at low temperatures.
Acknowledgments
We thank past and present members of the Cianciotto laboratory for their assistance and helpful comments. We also thank Cary Engleberg for the use of dot and lvh mutants.
This work was supported by NIH grant AI43987 awarded to N.P.C.
Footnotes
Published ahead of print on 28 December 2007.
REFERENCES
- 1.Basak, C., S. K. Pathak, A. Bhattacharyya, S. Pathak, J. Basu, and M. Kundu. 2005. The secreted peptidyl prolyl cis,trans-isomerase HP0175 of Helicobacter pylori induces apoptosis of gastric epithelial cells in a TLR4- and apoptosis signal-regulating kinase 1-dependent manner. J. Immunol. 174:5672-5680. [DOI] [PubMed] [Google Scholar]
- 2.Budde, I., L. Steil, C. Scharf, U. Volker, and E. Bremer. 2006. Adaptation of Bacillus subtilis to growth at low temperature: a combined transcriptomic and proteomic appraisal. Microbiology 152:831-853. [DOI] [PubMed] [Google Scholar]
- 3.Carvalho, F. R., R. F. Vazoller, A. S. Foronda, and V. H. Pellizari. 2007. Phylogenetic study of Legionella species in pristine and polluted aquatic samples from a tropical Atlantic forest ecosystem. Curr. Microbiol. 55:288-293. [DOI] [PubMed] [Google Scholar]
- 4.Cazalet, C., C. Rusniok, H. Bruggemann, N. Zidane, A. Magnier, L. Ma, M. Tichit, S. Jarraud, C. Bouchier, F. Vandenesch, F. Kunst, J. Etienne, P. Glaser, and C. Buchrieser. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36:1165-1173. [DOI] [PubMed] [Google Scholar]
- 5.Chakraborty, A., B. Sen, R. Datta, and A. K. Datta. 2004. Isomerase-independent chaperone function of cyclophilin ensures aggregation prevention of adenosine kinase both in vitro and under in vivo conditions. Biochemistry 43:11862-11872. [DOI] [PubMed] [Google Scholar]
- 6.Cianciotto, N. P. 2005. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13:581-588. [DOI] [PubMed] [Google Scholar]
- 7.Cianciotto, N. P., and B. S. Fields. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. USA 89:5188-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornelis, G. R. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811-825. [DOI] [PubMed] [Google Scholar]
- 9.DebRoy, S., V. Aragon, S. Kurtz, and N. P. Cianciotto. 2006. Legionella pneumophila Mip, a surface-exposed peptidylproline cis-trans-isomerase, promotes the presence of phospholipase C-like activity in culture supernatants. Infect. Immun. 74:5152-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DebRoy, S., J. Dao, M. Soderberg, O. Rossier, and N. P. Cianciotto. 2006. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc. Natl. Acad. Sci. USA 103:19146-19151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delepelaire, P. 2004. Type I secretion in gram-negative bacteria. Biochim. Biophys. Acta 1694:149-161. [DOI] [PubMed] [Google Scholar]
- 12.Donlan, R. M., T. Forster, R. Murga, E. Brown, C. Lucas, J. Carpenter, and B. Fields. 2005. Legionella pneumophila associated with the protozoan Hartmannella vermiformis in a model multi-species biofilm has reduced susceptibility to disinfectants. Biofouling 21:1-7. [DOI] [PubMed] [Google Scholar]
- 13.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 14.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fliermans, C. B., W. B. Cherry, L. H. Orrison, S. J. Smith, D. L. Tison, and D. H. Pope. 1981. Ecological distribution of Legionella pneumophila. Appl. Environ. Microbiol. 41:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardy, J. L., M. R. Laird, F. Chen, S. Rey, C. J. Walsh, M. Ester, and F. S. Brinkman. 2005. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21:617-623. [DOI] [PubMed] [Google Scholar]
- 17.Grindley, N. D., and C. M. Joyce. 1980. Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903. Proc. Natl. Acad. Sci. USA 77:7176-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hennecke, G., J. Nolte, R. Volkmer-Engert, J. Schneider-Mergener, and S. Behrens. 2005. The periplasmic chaperone SurA exploits two features characteristic of integral outer membrane proteins for selective substrate recognition. J. Biol. Chem. 280:23540-23548. [DOI] [PubMed] [Google Scholar]
- 20.Heuner, K., and M. Steinert. 2003. The flagellum of Legionella pneumophila and its link to the expression of the virulent phenotype. Int. J. Med. Microbiol. 293:133-143. [DOI] [PubMed] [Google Scholar]
- 21.Jacobi, S., and K. Heuner. 2003. Description of a putative type I secretion system in Legionella pneumophila. Int. J. Med. Microbiol. 293:349-358. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, T. L., J. Abendroth, W. G. Hol, and M. Sandkvist. 2006. Type II secretion: from structure to function. FEMS Microbiol. Lett. 255:175-186. [DOI] [PubMed] [Google Scholar]
- 23.Joly, J. R., M. Boissinot, J. Duchaine, M. Duval, J. Rafrafi, D. Ramsay, and R. Letarte. 1984. Ecological distribution of Legionellaceae in the Quebec City area. Can. J. Microbiol. 30:63-67. [DOI] [PubMed] [Google Scholar]
- 24.Mampel, J., T. Spirig, S. S. Weber, J. A. Haagensen, S. Molin, and H. Hilbi. 2006. Planktonic replication is essential for biofilm formation by Legionella pneumophila in a complex medium under static and dynamic flow conditions. Appl. Environ. Microbiol. 72:2885-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marrao, G., A. Verissimo, R. G. Bowker, and M. S. daCosta. 1993. Biofilms as major sources of Legionella spp. in hydrothermal areas and their dispersion into stream water. FEMS Microbiol. Ecol. 12:25-33. [Google Scholar]
- 26.Mouchtouri, V., E. Velonakis, A. Tsakalof, C. Kapoula, G. Goutziana, A. Vatopoulos, J. Kremastinou, and C. Hadjichristodoulou. 2007. Risk factors for contamination of hotel water distribution systems by Legionella species. Appl. Environ. Microbiol. 73:1489-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 28.O'Loughlin, R. E., L. Kightlinger, M. C. Werpy, E. Brown, V. Stevens, C. Hepper, T. Keane, R. F. Benson, B. S. Fields, and M. R. Moore. 2007. Restaurant outbreak of Legionnaires' disease associated with a decorative fountain: an environmental and case-control study. BMC Infect. Dis. 7:93-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paszko-Kolva, C., M. Shahamat, and R. R. Colwell. 1993. Effect of temperature on survival of Legionella pneumophila in the aquatic environment. Microb. Releases 2:73-79. [PubMed] [Google Scholar]
- 30.Pereira, P. J., M. C. Vega, E. Gonzalez-Rey, R. Fernandez-Carazo, S. Macedo-Ribeiro, F. X. Gomis-Ruth, A. Gonzalez, and M. Coll. 2002. Trypanosoma cruzi macrophage infectivity potentiator has a rotamase core and a highly exposed alpha-helix. EMBO Rep. 3:88-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piao, Z., C. C. Sze, O. Barysheva, K. Iida, and S. Yoshida. 2006. Temperature-regulated formation of mycelial mat-like biofilms by Legionella pneumophila. Appl. Environ. Microbiol. 72:1613-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pissavin, C., and N. Hugouvieux-Cotte-Pattat. 1997. Characterization of a periplasmic peptidyl-prolyl cis-trans isomerase in Erwinia chrysanthemi. FEMS Microbiol. Lett. 157:59-65. [DOI] [PubMed] [Google Scholar]
- 33.Ragull, S., M. Garcia-Nunez, M. L. Pedro-Botet, N. Sopena, M. Esteve, R. Montenegro, and M. Sabria. 2007. Legionella pneumophila in cooling towers: fluctuations in counts, determination of genetic variability by pulsed-field gel electrophoresis (PFGE), and persistence of PFGE patterns. Appl. Environ. Microbiol. 73:5382-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossier, O., and N. P. Cianciotto. 2005. The Legionella pneumophila tatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infect. Immun. 73:2020-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rossier, O., S. Starkenburg, and N. P. Cianciotto. 2004. Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires' disease pneumonia. Infect. Immun. 72:310-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saul, F. A., J. P. Arie, B. Vulliez-le Normand, R. Kahn, J. M. Betton, and G. A. Bentley. 2004. Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity. J. Mol. Biol. 335:595-608. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt, B., T. Tradler, J. U. Rahfeld, B. Ludwig, B. Jain, K. Mann, K. P. Rucknagel, B. Janowski, A. Schierhorn, G. Kullertz, J. Hacker, and G. Fischer. 1996. A cyclophilin-like peptidyl-prolyl cis/trans isomerase from Legionella pneumophila—characterization, molecular cloning and overexpression. Mol. Microbiol. 21:1147-1160. [DOI] [PubMed] [Google Scholar]
- 38.Scholz, C., B. Eckert, F. Hagn, P. Schaarschmidt, J. Balbach, and F. X. Schmid. 2006. SlyD proteins from different species exhibit high prolyl isomerase and chaperone activities. Biochemistry 45:20-33. [DOI] [PubMed] [Google Scholar]
- 39.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 40.Sheehan, K. B., J. M. Henson, and M. J. Ferris. 2005. Legionella species diversity in an acidic biofilm community in Yellowstone National Park. Appl. Environ. Microbiol. 71:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Söderberg, M. A., O. Rossier, and N. P. Cianciotto. 2004. The type II protein secretion system of Legionella pneumophila promotes growth at low temperatures. J. Bacteriol. 186:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinert, M., K. Heuner, C. Buchrieser, C. Albert-Weissenberger, and G. Glockner. 2007. Legionella pathogenicity: genome structure, regulatory networks and the host cell response. Int. J. Med. Microbiol. 297:577-587. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki, Y., M. Haruki, K. Takano, M. Morikawa, and S. Kanaya. 2004. Possible involvement of an FKBP family member protein from a psychrotrophic bacterium Shewanella sp. SIB1 in cold-adaptation. Eur. J. Biochem. 271:1372-1381. [DOI] [PubMed] [Google Scholar]
- 44.Wadowsky, R. M., R. Wolford, A. M. McNamara, and R. B. Yee. 1985. Effect of temperature, pH, and oxygen level on the multiplication of naturally occurring Legionella pneumophila in potable water. Appl. Environ. Microbiol. 49:1197-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wullings, B. A., and D. van der Kooij. 2006. Occurrence and genetic diversity of uncultured Legionella spp. in drinking water treated at temperatures below 15°C. Appl. Environ. Microbiol. 72:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]