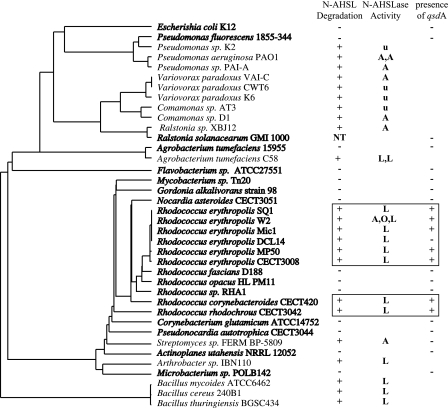
FIG. 5.
Phylogenetic distribution of N-AHSL degradation in Rhodococcus. The ability to degrade N-AHSLs among bacteria is highlighted on a phylogenetic tree based on the 16S rRNA gene. Strains shown in bold were tested in the present study. N-AHSL degradation data from the literature are reported for all other strains. Wherever possible the nature of the enzymatic activity is noted: A, N-AHSL amidohydrolase; L, N-AHSL lactonase; O, N-AHSL oxidoreductase; u, undetermined. The number of identified activities is also reported (e.g., A, A indicates two amidohydrolases). The presence (+) or absence (−) of a qsdA homologue was determined by hybridization with a qsdA probe. The N-AHSL-degrading strains belonging to the Rhodococcus genus are boxed. Agrobacterium tumefaciens strain C58 harbors two N-AHSL lactonases (AttM and AiiB), and R. erythropolis strain W2 harbors at least three N-AHSL modification/degradation enzymes: an amidohydrolase (A), an oxidoreductase (O), and a lactonase (L). NT, not tested.