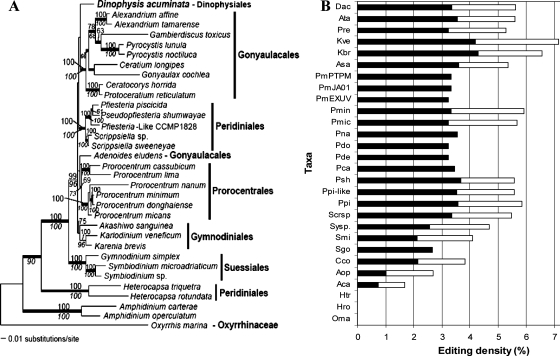
FIG. 4.
Maximum-likelihood tree inferred from 18S rDNA-cob-cox1 three-gene concatenated data set (A) and mapping of editing density (B). (A) The three-gene maximum-likelihood tree was inferred using PAUP with an ssGTR model (four categories of nucleotide substitution rates). Bootstrap support was assessed using PHYML with a GTR + I + Γ model (numbers above branches) and MP analysis (numbers below branches). Posterior probabilities from Bayesian analysis when ≥0.95 are indicated as thick branches. The tree is rooted with Oxyrrhis, the lineage that is consistently placed at the base of all dinoflagellates when trees are rooted with the apicomplexan Plasmodium. (B) Editing density is plotted against taxa (abbreviations of species names are as in Table 1), with filled bars of cob stacked by open bars of cox1. Note that in the phylogenetic tree, there is a well-supported early-to-later branching order from Oxyrrhis, to Amphidinium, to Heterocapsa, to Symbiodinium, to the clade of Akashiwo/Karlodinium/Karenia. Accordingly, editing density increases in this order. For the remaining lineages, phylogenetic resolution is less clear, and editing densities among those lineages are similar.