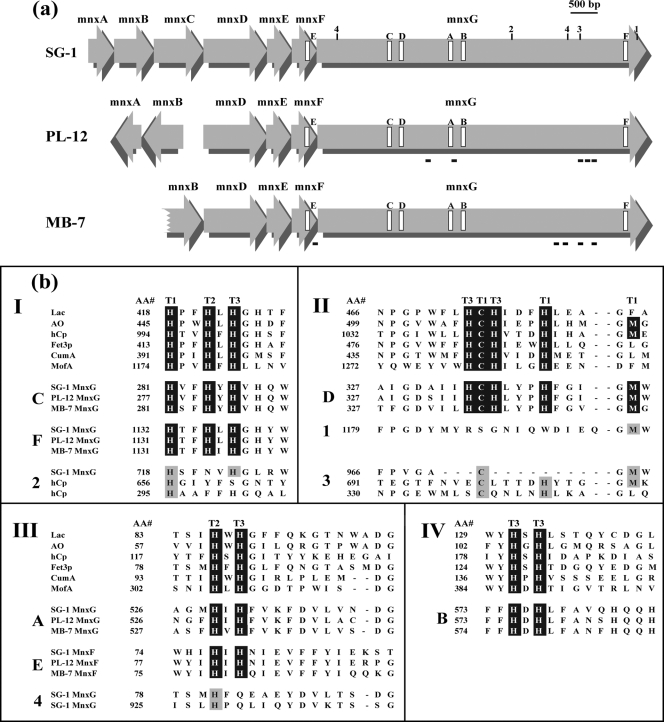
FIG. 3.
(a) Schematic representation of mnx region genes of Bacillus sp. strains SG-1, PL-12, and MB-7. The locations of putative Cu-binding regions are indicated by open rectangles and labeled A to F according to the data in panel b. Additional regions with potential Cu ligands are indicated by numbers (1-4) as in panel b. The locations of peptides detected by MS/MS analysis of the Mn oxide band from the in-gel activity assay are shown with black bars beneath the mnx genes of strains PL-12 and MB-7. (b) Alignment of putative mnx Cu-binding region amino acid sequences to consensus Cu-binding sequence motifs found in all MCOs. The consensus sequences shown (I to IV) are for the well-characterized MCOs laccase (Lac), ascorbate oxidase (AO), ceruloplasmin (hCp), and Fet3p, as well as putative MCOs involved in Mn(II) oxidation in P. putida (CumA) and L. discophora (MofA). “AA#” refers to the number of the first amino acid of each block. Note that the order of the MnxG Cu-binding regions (C, D, A, and B) is different than all other MCOs (III, IV, I, and II). Consensus MCO Cu-binding motifs are highlighted in black, and the type of Cu site for each ligand (based on characterized MCOs) is indicated with T1, T2, or T3. Mnx regions E and F are the fifth and sixth Cu-binding regions that are not found in other MCOs. Additional potential Cu-binding regions present in MnxG (only SG-1 shown here) are shown (sequences 1 to 4), with possible coordinating ligands highlighted with gray. Corresponding regions of hCp are also shown in sequences 2 and 3, with known Cu ligands highlighted in gray.