Abstract
The molecular structure and transferability of Tn1546 in 143 vancomycin-resistant Enterococcus faecium (VREF) isolates obtained from patients (n = 49), surface water (n = 28), and urban and hospital sewage (n = 66) in Tehran, Iran, were investigated. Molecular characterization of Tn1546 elements in vanA VREF was performed using a combination of restriction fragment length polymorphism analysis and DNA sequencing of the internal PCR fragments of vanA transposons. Long-PCR amplification showed that the molecular size of Tn1546 elements varied from 10.8 to 12.8 kb. The molecular analysis of Tn1546 showed that 45 isolates (31.5%) harbored a deletion/mutation upstream from nucleotide 170. No horizontal transfer of Tn1546 was observed following filter-mating conjugation with these isolates. Nevertheless, the rates of transferability for other isolates were 10−5 to 10−6 per donor. Insertion sequences IS1216V and IS1542 were present in 103 (72%) and 138 (96.5%) of the isolates, respectively. The molecular analysis of Tn1546 elements resulted in three genomic organizations. The genomic organization lineage 1 was dominated by the isolates from clinical samples (3.4%), lineage 2 was dominated mostly by sewage isolates (24.5%), and lineage 3 contained isolates obtained from all sources (72.1%). The genetic diversity determined using pulsed-field gel electrophoresis (PFGE) revealed a single E. faecium clone, designated 44, which was common to the samples obtained from clinical specimens and hospital and municipal sewage. Furthermore, the results suggest that lineage 3 Tn1546 was highly disseminated among our enterococcal isolates in different PFGE patterns.
Vancomycin and teicoplanin are glycopeptide antibiotics of clinical importance in the treatment of severe nosocomial infections caused by Staphylococcus and Enterococcus (5). Over the last 2 decades vancomycin-resistant enterococci have spread with unanticipated rapidity and have emerged as the major cause of nosocomial infections in clinical settings (3). The rapid rise in the antimicrobial resistance of enterococci has been attributed to clonal dissemination of the isolates and horizontal transfer of transposons containing vancomycin-resistant genes (12, 13, 17). It has been known that the vanA gene cluster is carried as a part of Tn1546-like elements, which in addition carry a cluster of nine genes mediating high-level of resistance to both vancomycin and teicoplanin (6). The nine polypeptides have been divided into four functional groups: transposition (ORF1 and ORF2), regulation (VanR and VanS), resistance to glycopeptides (VanH, VanA, and VanX), and synthesis of peptidoglycan (VanY and VanZ) (15).
In the present study the clonal variation of vancomycin-resistant Enterococcus faecalis (VREF) was investigated by pulsed-field gel electrophoresis (PFGE), and a detailed molecular analysis of Tn1546 elements was performed. The isolates were obtained from different sources in Tehran, Iran, which would give an overall circulation of VREF in human and water samples. Samples were taken from hospital and municipal sewage treatment plants, because they are favorable milieus, consisting of variable mixtures of bacteria, nutrients, and antimicrobial agents, for both survival and gene transfer (11). Enterococci were also isolated from the surface water, which could eventually infect humans (21). Molecular analysis of these isolates would therefore provide an understanding of the mechanism of the spread of vancomycin resistance and dissemination of VREF.
In view of the lack of information on the diversity of Tn1546 elements available in this region of the world, this study was undertaken to investigate the genetic relationship between vanA transposons of VREF obtained from different sources. The diversity of the Tn1546 elements was explored using long-PCR-restriction fragment length polymorphism (L-PCR-RFLP), PCR analysis of internal regions, and Southern hybridization. Conjugation experiments were done to assess the mobilities of various Tn1546 elements among our enterococcal isolates.
MATERIALS AND METHODS
VREF sample collection.
All VREF isolates were collected during 2006. The clinical samples (n = 49) were collected from hospitalized patients in three major hospitals in Tehran, Iran. The samples were urine, wound specimens, blood, body fluids, respiratory tract specimens, and abscesses. The isolation of VREF from clinical specimens was carried out by standard microbiological techniques. Gram-positive cocci were tentatively identified as enterococcus on the basis of negative catalase test, growth in 6.5% sodium chloride, hydrolysis of esculin, and pyrrolidonyle arylamidase test. The susceptibility to vancomycin was determined by the disc diffusion method and MIC determination with Etest. The VREF environmental samples were collected from three urban sewage treatment plants, two hospital sewage sites, and surface water located in different parts of Tehran. Isolation of the VREF isolates from sewage treatment plants was done as described previously (18). In brief, sewage treatment plant samples were cultured on mEnterococcus agar (Becton Dickinson and Co., Sparks, MD) plates supplemented with 8 μg of vancomycin per ml. Representative isolates were collected and typed using the PhPlate-RF plates (PhPlate AB, Stockholm, Sweden) as described in detail previously (7, 18). Representative isolates were kept for further studies. Enterococcal species identification was performed by using conventional tests, including 0.04% telurite reduction, carbohydrate utilization, arginine dehydrolase activity, methyl-α-d-glucopyranoside acidification, motility, pigmentation, and PCR using species-specific primers (8). E. faecium BM4147 and E. faecalis V583 were used as positive controls and were obtained from the Pasteur Institute, Paris.
PCR and RFLP.
Extraction of bacterial DNA was performed with a DNeasy kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. The vancomycin resistance genotype was determined by PCR using primers specific for vanA (10). The extracted DNA was quantified spectrophotometrically, and 125 ng was used as templates for L-PCRs. Initially, DNA was amplified with the P1 primer, an oligonucleotide complementary to the inverted repeats flanking Tn1546, using the Expand long-template PCR system (Roche Diagnostic GmbH, Mannheim, Germany). For isolates that did not amplify with P1, the amplification was subsequently performed using primers specific for orf2 forward and P1. Amplification was done with the following protocol: initial denaturation at 94°C for 2 min; 10 cycles of denaturation at 95°C for 15 s, annealing at 62°C for 30 s, and extension at 68°C for 8 min; 20 cycles of denaturation at 95°C for 20 s, annealing at 65°C for 30 s, and extension at 68°C for 8 min (with the elongation time increased by 20 s per cycle); and a final extension at 68°C for 7 min. All PCR was done using E. faecium BM4147 as the prototype. Following L-PCR amplification, enzymatic digestion of the products was done with 30 U of ClaI (Roche Diagnostic GmbH).
PCR amplification of internal regions of Tn1546 was also performed with the primers orf1 (orf1-1 to orf1-6), orf2-F, orf2-R, vanR-F, vanS-F, vanS-R, vanH1, vanX2, vanY1, vanY2, vanZ1 and vanZ2, as listed in Table 1. The PCR amplicons of the internal region of the Tn1546-like elements were produced by an initial cycle at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 3.5 min and a final extension at 72°C for 10 min.
TABLE 1.
Primers used in this study
| Gene | Sequence | Attachment site(s) | Reference |
|---|---|---|---|
| P1 | 5′-GGAAAATGCGGATTTACAACGCTAAG | 13-38, 10840-10814 | 23 |
| orf1-1 | 5′-AACCTAAGGGCGACATATGGT | 164-185 | 6 |
| orf1-2 | 5′-AGGGCGACATATGGTGTAACA | 170-190 | 6 |
| orf1-3 | 5′-AAAAGGAGCCACCATCTACCG | 921-901 | 6 |
| orf1-4 | 5′-GCATGTAGTGATGAAACACCTAGCTGC | 949-975 | 6 |
| orf1-5 | 5′-CGTCCTGCCGACTATGATTATTT | 1913-1891 | 6 |
| orf1-6 | 5′-TGAAGATGAATGGATACTGGGGACC | 2976-2952 | 6 |
| orf2-f | 5′-TCATTCCATTTCTGTATTTTCAATTT | 3048-3073 | This study |
| orf2-f | 5′-ACTAATGTATCTAGGGCTTCA | 3709-3729 | This study |
| orf2-r | 5′-GCCCATTAGCGGAATACAGA | 3786-3767 | This study |
| vanH1 | 5′-ATGAATAACATCGGCATTAC | 6018-637 | 23 |
| vanX1 | 5′-ATGGAAATAGGATTTACTTT | 8016-8035 | 23 |
| vanX2 | 5′-TTATTTAACGGGGAAATC | 8624-8607 | 23 |
| vanR | 5′-AGACAAGTCTGAGATTGACCTTGCC | 4101-4125 | 4 |
| vanS | 5′-AACGACTATTCCAAACTAGAA | 4676-4696 | 6 |
| vanS-r | 5′-GCTGGAAGCTCTACCCTAAA | 5769-5750 | 6 |
| vanR-r | 5′-GCAATTTCATGTTCATCATCCA | 4018-3998 | This study |
| vanX-f | 5′-ATGGGTATTTTCAGAAGTCCC | 9212-9193 | This study |
| vanY1 | 5′-AGAGACGAACCATACCCCAA | 8577-8596 | 9 |
| vanZ1 | 5′-CTGGGAATTTCAGAGAGATG | 10258-10277 | 9 |
| vanZ2 | 5′-AATGGGTACGGTAAACGAGC | 10581-10562 | 9 |
| vanY2 | 5′-GTTTCCCGGATCAACACATACTA | 9927-9949 | 9 |
Presence of IS.
A combination of RFLP analysis and DNA sequencing of the internal PCR fragments of VanA transposons was used for determination of the locations of insertion sequences (IS). Specific primers targeting the IS elements within Tn1546 were designed (Table 1).
DNA sequencing.
PCR products of internal regions in vanR, vanS, vanH, vanA, vanXYZ, and IS of Tn1546 were cloned in the Promega TA Easy vector (Madison, WI), and sequencing was carried out using the ABI capillary system (Macrogen Research, Seoul, Korea).
PFGE and Southern hybridization.
PFGE was performed on a CHEF-DR III apparatus (Bio-Rad Laboratories, Richmond, CA) as described previously (19). After digestion with SmaI, genomic DNA was separated by electrophoresis, with ramped pulse times beginning with 5 s and ending with 35 s at 6 V/cm for 27 h. The banding patterns were interpreted by Dice analysis and clustered by the unweighted pair group method with arithmetic averages with Gelcompar II version 4.0 (Applied Maths, Sint-Matens-Latem, Belgium). Southern blot hybridization was performed using a digoxigenin-labeled vanA probe (9).
Conjugation experiments.
Cross-streak testing of VREF was performed with E. faecalis JH2-2 and E. faecium 4107, separately. For the isolates with positive results, E. faecalis strain JH2-2 was used as the recipient in the conjugation assay. Filter mating was performed using a 1:1 donor-recipient mixture. After incubation at 37°C for 24 h, transconjugants were selected on brain heart infusion agar containing vancomycin (20 mg/ml), rifampin (20 mg/ml), and fusidic acid (10 mg/ml). The transconjugants were analyzed by antibiotic sensitivity assay and PCR for species identification, the presence of the vanA gene, and transfer of the complete transposon (2).
RESULTS
Genetic organizations of Tn1546-like elements.
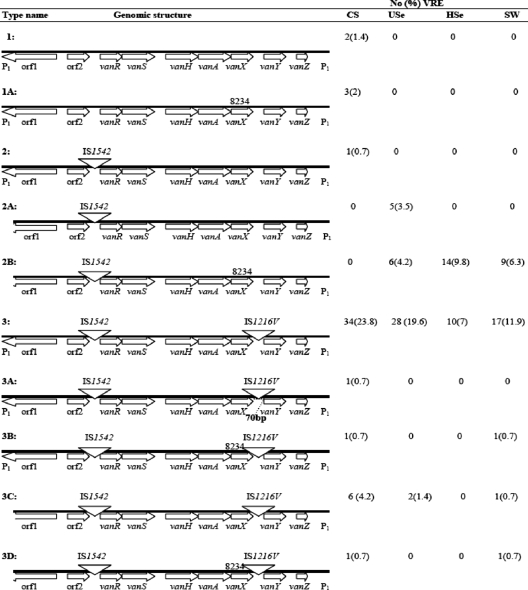
Three characteristics, i.e., point mutations, IS elements, and gene deletions were considered when examining the genomic organizations of 143 E. faecium isolates. The results revealed the presence of three major genomic lineages of Tn1546, designated 1 to 3, and seven subtypes. Each of the three lineages contained between one and four subtypes. The MICs of vancomycin and teicoplanin for all isolates were ≥128 to 256 μg/ml and ≥8 to 256 μg/ml, respectively.
The complete Tn1546, with no deletions, was detected in 98 isolates (68%) within all lineages. Of these, 42 (29%) and 56 (39%) were isolated from patients and water samples (hospital and urban sewage and surface water), respectively. The results showed that lineage 1 contained the isolates from the hospitalized patients only.
The majority of isolates within lineage 2 were from the sewage and surface water samples (Fig. 1). Tn1546 of lineage 3 and subtype 2B constituted 92% of the total isolates which were collected from all sources. Lineage 1 of Tn1546 was found to be identical to the prototype BM4147 (Fig. 1). Lineage 1, with 5 isolates (3.4%), was found only in the clinical isolates, suggesting that 96.5% of the isolates carried Tn1546 which was different from that of the prototype BM4147 strain.
FIG. 1.
Schematic genomic organizations of Tn1546 and prevalence of each pattern among 143 E. faecium isolates obtained from different sources. The isolates were obtained from patients (CS), hospital (Hse) and urban (Use) sewage, and surface water (SW).
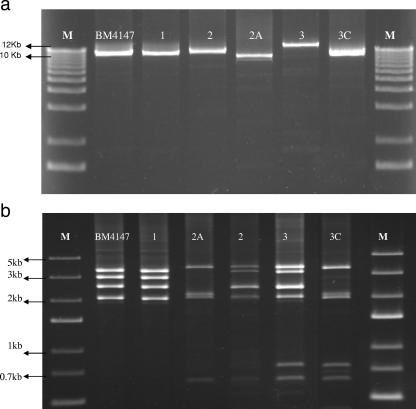
L-PCR-RFLP of Tn1546 with ClaI for lineage 2 was characterized by an additional band of about 600 bp in comparison to lineage 1 (Fig. 2b). Moreover, lineage 3 was found to carry two additional bands of about 700 and 600 bp. L-PCR-RFLP of Tn1546 with a single mutation at position 8234 (subtypes 1A, 2B, 3B, and 3D) or a deletion in the intergenic region of vanX (3A) was similar to that for the subtypes with no mutation or deletion.
FIG. 2.
(a) Amplification products of L-PCR of different types of transposons. (b) Resulting L-PCR-RFLP products using the ClaI restriction enzyme.
The amplification of internal regions and the designing three new primers in this study revealed that lineages 2 and 3 harbored an additional 1.3 kb (Fig. 1) which positioned in the orf2-vanR region (Fig. 1). Furthermore, the results showed that lineage 3 carried an extra 0.7-kb fragment in the vanX-vanY region. The additional fragment was found to be insertional element. The results of sequencing analysis revealed that the IS elements in the orf2-vanR and vanX-vanY regions were of the IS1542 (1.3 kb) and IS1216V (0.7 kb) types, respectively (Fig. 1). IS1542 elements were detected in 138 isolates (96.5%), while IS1216V elements were detected in 103 isolates (72%). No IS was found in five (3.4%) of the isolates. All IS1216V and IS1542 elements were of the expected sizes and oriented in the opposite direction. IS1216V was found to be inserted at position 8731 between vanX and vanY, and IS1542 positioned at nucleotide 3932 (Fig. 1). Furthermore, sequencing showed the presence of a point mutation at positions 490 (A→T) and 664 (G→A) based on the sequence of IS1216V (GenBank accession no. AF093508.1). All VREF strains were analyzed for the presence of a single mutation at vanX position 8234 by PCR-RFLP with the DdeI restriction enzyme and examined by sequencing. Of 143 isolates, 36 (25%) carried a mutation in subtypes 1A, 2B, 3B, and 3D of Tn1546. Among the genomic arrangements of Tn1546, only subtype 3A showed a 72-nucleotide deletion segment which was associated with IS1216V downstream of vanX. Subtype 3A was found in 1 isolate (0.7%) only.
A unique characteristic of all of our isolates was the presence of a single point mutation at position 5727 in the vanS element.
PFGE and DNA hybridization.
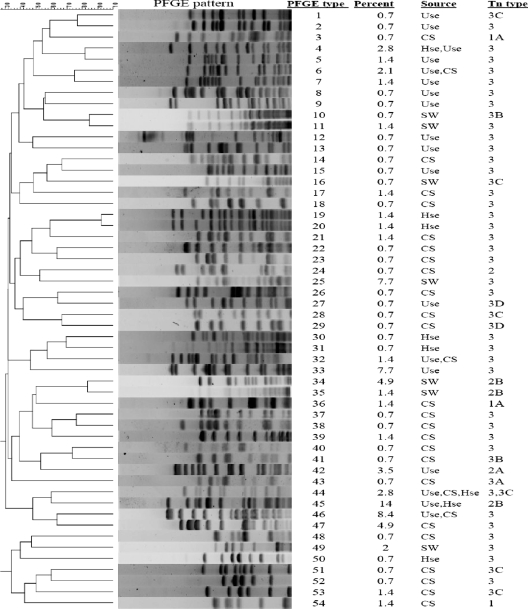
PFGE analysis of the 143 isolates showed that they were extremely heterogeneous. Fifty-four PFGE types were identified, with 24 types constituting 113 identical isolates with more than 95% similarity. The remaining 30 isolates (29.6%) were highly diverse, belonging to 30 PFGE types. All clinical isolates (49) were classified into 20 distinct PFGE types and 9 common types. The isolates from surface water (n = 28) and from hospital (n = 24) and municipal (n = 42) sewage were grouped in 8, 9, and 18 PFGE types, respectively. PFGE showed that some isolates were common among samples taken from clinical specimens and from hospital and municipal sewage (Fig. 3). The representative PFGE patterns are shown in Fig. 3.
FIG. 3.
Unweighted pair group method with arithmetic averages dendrogram of the representative PFGE patterns of isolates obtained from different sources. PFGE patterns (n = 54) of the isolates obtained from patients (CS), hospital (Hse) and urban (Use) sewage, and surface water (SW) are shown, as are the percentages of the pattern found among the total of 143 VREF isolates.
Southern blot hybridization analysis of the isolates showed that the isolates carried the vanA gene residing on a 78-kb DNA fragment. The only exception was that one isolate contained the vanA gene residing on a DNA fragment of 170 kb.
Conjugation analysis.
Filter-mating conjugation assay revealed that most VREF isolates could transfer the vancomycin-resistant elements to E. faecalis JH2-2. The efficiency of conjugation ranged from 10−5 to 10−10. The conjugation rate was significantly higher in isolates with Tn1546 with lineage 3. The antibiotic profile, vanA gene, and species in the transconjugants were examined, and it was shown that complete transfer of Tn1546 to the recipient cells (E. faecalis JH2-2) has occurred. No transfer of vancomycin resistance was seen with the isolates harboring a deletion/mutation upstream of nucleotide 170 of Tn1546. This was seen with the isolates containing Tn1546 with the genomic arrangements 2A, 2B, 3C, and 3D.
DISCUSSION
Molecular characterizations of Tn1546 have been fully elucidated and show limited diversity (4, 6, 9). Moreover, it is important to understand the dissemination of Tn1546 subtypes among the enterococcal species. In the present study, we identified and characterized polymorphic regions of Tn1546-like elements of 143 VREF strains isolated from different sources. Detailed molecular characterization was performed by a combination of RFLP analysis and DNA sequencing of the internal PCR fragments of VanA transposons. In general, our results showed point mutations, insertions of IS elements, and deletions in Tn1546 elements among the isolates.
Overall, three lineages and seven subtypes of Tn1546 elements could be distinguished in our isolates. A scheme was constructed to elucidate the relationship between Tn1546 variants found in this study. Similar to the report by Arthur and colleagues (1), we showed that Tn1546 element lineage 1 was similar to prototype BM4147, which constituted only a small portion of our isolates.
It has been suggested that IS1216V is ubiquitous in the vanA elements (9). In this study, we found that 72% of our isolates obtained from all sources carried IS1216V. The presence of IS1216V has also been reported in majority of isolates obtained from Europe and North America (9, 17), suggesting a global distribution of this IS. Furthermore, this IS has been reported in other species, such as Staphylococcus aureus (14).
On the other hand, there are only few published reports about the distribution of IS1542, which suggests that this IS is geographically limited. The enterococcal isolates carrying IS1542 have been reported in Europe (22). To our knowledge, no reports have been published from North America in regard to circulation of IS1542 in enterococcal populations (17). We were also able to detect IS1542 in the majority of our isolates (96%) originating from patients as well as surface water and hospital and municipal sewage, suggesting widespread dissemination of IS1542 among E. faecium isolates in Iran and also sharing of the common IS1542 between Europe and Iran.
Similar to the case for other studies (6), we also found that IS1542 and IS1216V elements were positioned at the left and right ends of transposons, respectively. The two IS examined here were found to be positioned reversely for all isolates which is commonly reported in Europe (20). Among all the isolates, subtype 3A Tn1546 was found to have a deletion of 73 nucleotides in the IS1216V integration site at position 8731. In contrast to another report (6), we found this deletion in only 0.7% (1) of the isolates.
About 30.2% of the Tn1546 transposons, including subtypes 2A (3.5%), 2B (20%), 3C (6.3%), and 3D (1.4%), could not be amplified using P1 primers. Further studies showed the possible occurrence of a deletion and/or mutation upstream from nucleotide 170. This mutation has been reported commonly elsewhere (16). It has been reported that deletions in the transposase and resolvase regions can abolish the transposition, which, in turn, may affect the dissemination of Tn1546-like elements (20). This was also supported by the results from our conjugation assay, where the isolates with 2B, 3C, and 3D Tn1546 either failed to transfer or showed a very low rate of transfer (i.e., 10−10) of vanA to the recipient bacteria. Moreover, the vanA gene transfer rates of Tn1546 containing the P1 region, such as 1A or 3A, was about 10−5 to 10−6 per donor, suggesting that the transposonal integrity would facilitate the horizontal transfer of resistance genes. The results showed that similar conjugation rates of vanA were obtained when isolates with the same subtype of Tn1546 were collected, regardless of the source.
In contrast to the report by Willems and colleagues (20), who have shown the presence of a point mutation at position 826 of IS1216V, we could observe the point mutations only at positions 490 (A→T) and 664 (G→A) of IS1216V. These mutations were found in only one isolate, which also showed a unique PFGE pattern (data not shown). Moreover, we found a point mutation in the vanS gene at position 5727 (C→A), resulting in change from glutamic acid to valine. Such a unique mutation in Tn1546 was found in all Iranian isolates regardless of their source of isolation, subtype, and range of MIC to vancomycin and teicoplanin.
The analysis of PFGE patterns exhibited high diversity in our VREF isolates, with the majority of them showing distinct PFGE patterns. Some PFGE patterns were common among isolates from patients and those obtained from other sources. The most common PFGE pattern was found to be the pattern designated 45, with 20 (14%) isolates. This genotype of E. faecium was found in hospital and municipal sewage, and all of these strains carried Tn1546 subtype 2B. In the absence of transfer of Tn1546 in the isolates with PFGE pattern 45, we postulate that the dispersion of these VREF isolates is through clonal dissemination and not horizontal transfer of vancomycin resistance gene. On the other hand, the PFGE patterns designated 6 and 32 contained isolates found in the clinical and urban sewage samples and carried lineage 3 of Tn1546, with a conjugation rate of 10−5. It is interesting to note that these isolates were collected at different time intervals, suggesting that the horizontal vanA transfer among this group of E. faecium strains could be the reason for dissemination of vancomycin resistance in enterococcal populations. In contrast, the PFGE pattern designated 44 contained the isolates obtained from clinical isolates and urban and hospital sewage, indicative of the clonal dissemination of this VREF clone (Fig. 3).
Among all isolates, lineage 3 of Tn1546 was the predominant lineage, accounting for 62% of the VREF isolates. This lineage could be found in the majority of isolates from patients (69%), urban sewage (67%), hospital sewage (42%), and surface water (61%). Such a large dissemination of lineage 3 Tn1546 was, in part, due to high rate of conjugation (10−5) of this particular transposon.
By Southern blot hybridization with a vanA-specific probe to DNA fragments from PFGE of SmaI-digested total DNA, Tn1546 was detected at one position. With the exception of one isolate which showed Tn1546 residing in a 170-kb DNA fragment, the isolates showed integration of Tn1546 in a 78-kb DNA fragment. Moreover, the conjugation assay showed that the strain with Tn1546 located in the 170-kb DNA fragment could not transfer the vancomycin resistant determinant to the recipient.
In conclusion, we found diverse Tn1546 elements among a large number of VREF isolates in this study. The presence of predominant lineage 3 Tn1546 may suggest that these elements can disseminate rapidly and acquire different genomic organizations in enterococcal populations. Furthermore, we observed that many of the molecular characteristics of VREF in our isolates were similar to those of isolates reported form Europe.
Acknowledgments
We thank Patrice Courvalin from the Pasteur Institute, Paris, France, for providing E. faecalis V583 with vanB and E. faecium BM4147 with vanA.
This work was supported in part by the World Health Organization, Eastern Mediterranean Regional Office, grant no. R6/18/3; Swedish International Development Cooperation Agency (Sida) grant no. 6342; and the Ministry of Health of Iran, Undersecretariat of Research.
Footnotes
Published ahead of print on 11 January 2008.
REFERENCES
- 1.Arthur, M., C. Molinas, F. Depardieu, and P. Courvalin. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clewell, D. B. 1981. Plasmids, drug resistance, and gene transfer in genus transfer in genus Streptococcus. Microbiol. Rev. 45:409-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eom, J. S., I. S. Hwang, B. Y. Hwang, J. G. Lee, Y. J. Lee, H. J. Cheong, Y. H. Park, S. C. Park, and W. J. Kim. 2004. Emergence of vanA genotype vancomycin-resistant enterococci with low or moderate levels of teicoplanin resistance in Korea. J. Clin. Microbiol. 42:1785-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garnier, F., K. Gambarotto, F. Denis, and M. C. Ploy. 2004. Molecular study of vancomycin-resistant enterococci isolated from humans and from food in a cattle-rearing area of France. J. Antimicrob. Chemother. 54:236-239. [DOI] [PubMed] [Google Scholar]
- 5.Hasman, H., F. M. Aarestrup, A. Dalsgaard, and L. Guardabassi. 2006. Heterologous expression of glycopeptide resistance vanHAX gene clusters from soil bacteria in Enterococcus faecalis. J. Antimicrob. Chemother. 57:648-653. [DOI] [PubMed] [Google Scholar]
- 6.Huh, J. Y., W. G. Lee, K. Lee, W. S. Shin, and J. H. Yoo. 2004. Distribution of insertion sequences associated with Tn1546-like elements among Enterococcus faecium isolates from patients in Korea. J. Clin. Microbiol. 42:1897-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iversen, A., I. Kühn, M. Rahman, A. Franklin, L. G. Burman, B. Olsson-Liljequist, E. Torell, and R. Möllby. 2004. Evidence for transmission between humans and the environment of a nosocomial strain of Enterococcus faecium. Environ. Microbiol. 1:55-61. [DOI] [PubMed] [Google Scholar]
- 8.Jackson, C. R., P. J. Fedorka-Cray, and J. B. Barrett. 2004. Use of genus- and species-specific multiplex PCR for identification of enterococci. J. Clin. Microbiol. 42:3558-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen, L. B., P. Ahrens, L. Dons, R. N. Jones, A. M. Hammerum, and F. M. Aarestrup. 1998. Molecular analysis of Tn1546 in Enterococcus faecium isolated from animals and humans. J. Clin. Microbiol. 36:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kariyama, R., R. Mitsuhata, J. W. Chow, D. B. Clewell, and H. Kumon. 2000. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J. Clin. Microbiol. 38:3092-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindberg, R., P. Jarnheimer, B. Olsen, M. Johansson, and M. Tysklind. 2004. Determination of antibiotic substances in hospital sewage water using solid phase extraction and liquid chromatography/mass spectrometry and group analogue internal standards. Chemosphere 57:1479-1488. [DOI] [PubMed] [Google Scholar]
- 12.Nichol, K. A., M. Sill, N. M. Laing, J. L. Johnson, D. J. Hoban, and G. G. Zhanel. 2006. Molecular epidemiology of urinary tract isolates of vancomycin-resistant Enterococcus faecium from North America. Int. J. Antimicrob. Agents 27:392-396. [DOI] [PubMed] [Google Scholar]
- 13.Novais, C., T. M. Coque, J. C. Sousa, F. Baquero, and L. Peixe. 2004. Local genetic patterns within a vancomycin-resistant Enterococcus faecalis clone isolated in three hospitals in Portugal. Antimicrob. Agents Chemother. 48:3613-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Périchon, B., and P. Courvalin. 2006. Synergism between beta-lactams and glycopeptides against VanA-type methicillin-resistant Staphylococcus aureus and heterologous expression of the vanA operon. Antimicrob. Agents Chemother. 50:3622-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ribeiro, T., M. Abrantes, M. D. F. Silva Lopes, and M. T. Barreto Crespo. 2007. Vancomycin-susceptible dairy and clinical enterococcal isolates carry vanA and vanB genes. Int. J. Food Microbiol. 113:289-295. [DOI] [PubMed] [Google Scholar]
- 16.Schouten, M. A., R. J. Willems, W. A. Kraak, J. Top, J. A. Hoogkamp-Korstanje, and A. Voss. 2001. Molecular analysis of Tn1546-like elements in vancomycin-resistant enterococci isolated from patients in Europe shows geographic transposon-type clustering. Antimicrob. Agents Chemother. 45:986-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simjee, S., D. G. White, P. F. McDermott, D. D. Wagner, M. J. Zervos, S. M. Donabedian, L. L. English, J. R. Hayes, and R. D. Walker. 2002. Characterization of Tn1546 in vancomycin-resistant Enterococcus faecium isolated from canine urinary tract infections: evidence of gene exchange between human and animal enterococci. J. Clin. Microbiol. 40:4659-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talebi, M., F. Rahimi, M. Katouli, I. Kühn, R. Möllby, S. Eshraghi, and M. R. Pourshafie. 2007. Prevalence and antimicrobial resistance of enterococcal species in sewage treatment plants in Iran. Water Air Soil Pollut. 185:111-119. [Google Scholar]
- 19.Turabelidze, D., M. Kotetishvili, A. Kreger, J. G. Morris, and A. Sulakvelidze. 2000. Improved pulsed-field gel electrophoresis for typing vancomycin-resistant enterococci. J. Clin. Microbiol. 38:4242-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willems, R. J. L., T. Janetta, N. Braak, A. Belkum, D. J. Mevius, G. Hendriks, M. Santen-Verheuvel, and J. D. A. Embden. 1999. Molecular diversity and evolutionary relationships of Tn1546-like elements in enterococci from humans and animals. Antimicrob. Agents Chemother. 43:483-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Witte, W. 2000. Ecological impact of antibiotic use in animals on different complex microflora: environment. Int. J. Antimicrob. Agents 14:321-325. [DOI] [PubMed] [Google Scholar]
- 22.Woodford, N., A. M. A. Adebiyi, M. I. Palepou, and B. D. Cookson. 1998. Diversity of VanA glycopeptide resistance elements in enterococci from human and nonhuman sources. Antimicrob. Agents Chemother. 42:502-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu, H. S., S. Y. Seol, and D. T. Cho. 2003. Diversity of Tn1546-like elements in vancomycin-resistant elements isolated from humans and poultry in Korea. J. Clin. Microbiol. 41:2641-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]