FIG. 2.
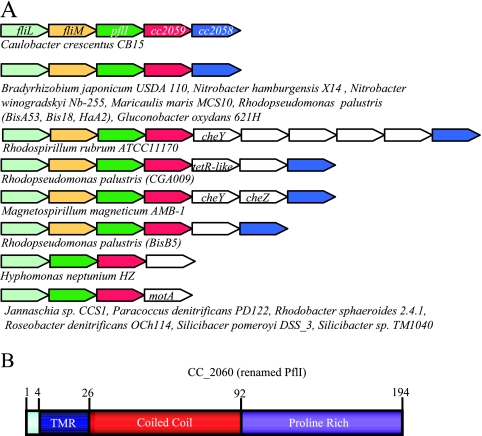
The three genes identified as candidates for a putative role in polar localization of flagella are found in similar chromosomal loci. (A) Genome regions of candidate genes. There are three genes that are largely present in polarly flagellated alphaproteobacteria (including C. crescentus) and absent from all other types of bacteria. Shown here are schematic diagrams of the genome regions of these genes. Homologs of fliL, fliM, CC_2060, CC_2059, and CC_2058 (pflI) are shown in light green, yellow, green, red, and blue, respectively. Other genes are shown in white. The directions of the arrows indicate the relative directions of transcription. The organisms listed under each schematic are the species with that particular genome organization. (B) Domain architecture of PflI. In silico analysis of PflI reveals three domains: a transmembrane region from residues 4 through 25 (TMR) shown in blue, a coiled-coil region from residues 26 through 92 shown in red, and an uncharacterized proline-rich region from residues 93 through 194, in which 24 of 102 amino acids are prolines, shown in purple.