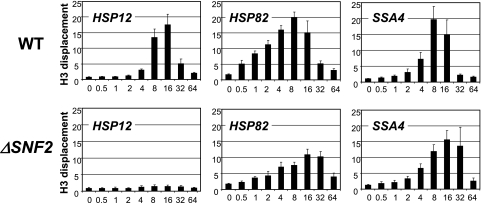
FIG. 2.
Chromatin remodeling is eliminated at the HSP12 promoter and delayed at the HSP82 and SSA4 promoters in the strain lacking SNF2. ChIPs using an antibody raised against the C terminus of histone H3 were performed with the ΔSNF2 (FY1360) or isogenic WT strain (FY1350). The promoter analyzed is indicated in the upper left corner of each panel. y axis, n-fold displacement of histone H3; x axis, time after heat shock (0 to 64 min). All real-time PCR values were normalized relative to the input and to the signal from the PHO5 promoter, which is known to contain positioned nucleosomes that do not change during heat shock (16). Values represent means ± standard deviations (n ≥ 3). Note: displacement value is an inverse value of abundance.