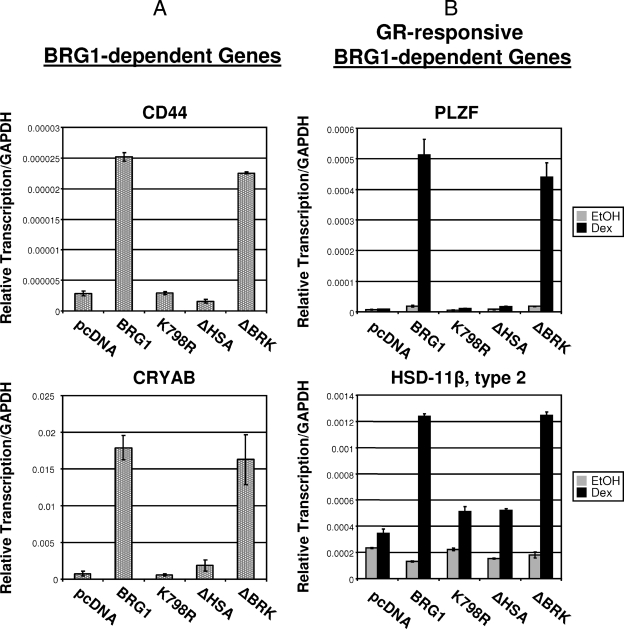
FIG. 4.
Comparison of the abilities of BRG1, ΔHSA, and ΔBRK to activate expression of endogenous BRG1-dependent or GR-responsive BRG1-dependent genes. (A) Real-time reverse transcription-PCR analysis was used to determine the expression level of BRG1-dependent genes, CD44 or CRYAB, in SW-13 wild-type cells expressing BRG1 or mutant proteins. (B) Reverse transcription-PCR expression analysis of GR-responsive BRG1-dependent genes, PLZF or HSD-11β type 2, in treated SW-13 cells expressing GR and BRG1 or mutant proteins. Equal amounts of cDNA were used in real-time PCRs with primers specific for endogenous genes CD44, CRYAB, PLZF, or HSD-11β type 2. Quantitative analysis was performed using Mx3000P software. Data are normalized to GAPDH and displayed as relative transcription. Values are shown as means plus standard deviations (error bars) from three experiments. EtOH, ethanol.