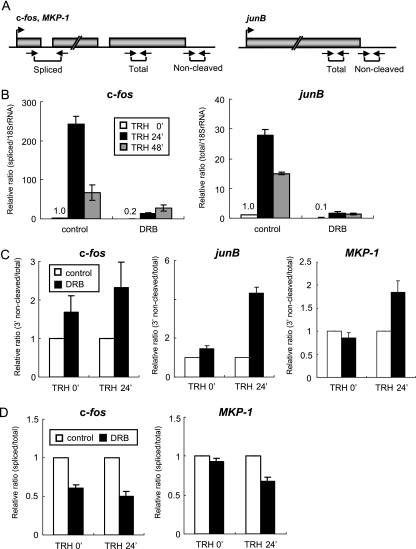
FIG. 3.
DRB suppresses IEG transcription and transcript processing. (A) Schematic diagrams, including exons and introns, with the locations of the primer sets used in the experiments. The primer sets for amplification of objective transcripts were designed based on the genetic information (c-fos, GenBank X06769; junB, GenBank NM_021836; MKP-1, GenBank NM_053769). (B) Relative amounts of c-fos and junB transcripts quantified by real-time RT-PCR (normalized to 18S RNA). (C) PCR-based analysis of the 3′-end cleavage ratio. Transcripts of the c-fos, junB, and MKP-1 genes were amplified with primer sets for detection of 3′-uncleaved and total RNAs. The values of 3′-uncleaved RNAs normalized to total RNAs are shown as relative 3′-end noncleavage ratios. (D) PCR-based analysis of the splicing ratio. c-fos and MKP-1 transcripts were amplified with primer sets for detection of spliced versus total RNA. The value of spliced RNA normalized to total RNA are shown as relative splicing ratios. (B to D) Each experiment was performed three times, and the relative ratio is shown as the mean ± standard error of the mean.