Abstract
The commensal microbiota protects the murine host from enteric pathogens. Nevertheless, specific pathogens are able to colonize the intestinal tract and invade, despite the presence of an intact biota. Possibly, effective pathogens disrupt the indigenous microbiota, either directly through pathogen-commensal interaction, indirectly via the host mucosal immune response to the pathogen, or by a combination of these factors. This study investigates the effect of peroral Salmonella enterica serovar Typhimurium infection on the intestinal microbiota. Since the majority of the intestinal microbiota cannot be cultured by conventional techniques, molecular approaches using 16S rRNA sequences were applied. Several major bacterial groups were assayed using quantitative PCR. Administration of either the 50% lethal dose (LD50) or 10× LD50 of Salmonella enterica serovar Typhimurium caused changes in the microbiota throughout the intestinal tract over the time course of infection. A 95% decrease in total bacterial number was noted in the cecum and large intestine with 10× LD50 S. enterica serovar Typhimurium challenge at 7 days postinfection, concurrent with gross evidence of diarrhea. In addition, alterations in microbiota composition preceded the onset of diarrhea, suggesting the involvement of pathogen-commensal interactions and/or host responses unrelated to diarrhea. Microbiota alterations were not permanent and reverted to the microbiota of uninfected mice by 1 month postinfection. Infection with a Salmonella pathogenicity island 1 (SPI1) mutant did not result in microbiota alterations, while SPI2 mutant infections triggered partial changes. Neither mutant was capable of prolonged colonization or induction of mucosal inflammation. These data suggest that several Salmonella virulence factors, particularly those involved in the local mucosal host response, are required for disruption of the intestinal ecosystem.
The intestinal microbial ecosystem, denoted the intestinal microbiota, has been implicated in normal intestinal growth and development, vitamin synthesis, fermentation of nondigestible food to extract nutrients, and mucosal protection (14, 23). Mucosal protection, sometimes referred to as “colonization resistance,” is thought to be mediated by the biota through monopolization of mucus attachment sites, competition for nutrients, and fierce niche protection by production and secretion of bacteriocins (23). The significance of the protective role of the microbiota has been highlighted by the profound impact seen when the microbiota is absent or disrupted (22, 25). Germfree mice have poorly developed mucosal architecture and rudimentary development of the mucosa-associated lymphoid tissue, are generally small and underweight, and are unable to digest their intestinal mucus (35). Germfree mice are also highly susceptible to intestinal infection. Pretreatment of mice with antibiotics, such as streptomycin, renders mice more susceptible to intestinal infection as well (7, 24). Antibiotic treatment appears to disrupt or eliminate the microbial colonization of the gut (33), although the effects of specific antibiotics on the microbiota have not been well characterized using culture-independent methods. The intestinal microbiota is a complex ecosystem made up of 500 to 1,000 different bacterial species (11), the majority of which are unculturable. Recent advances in molecular approaches to identification and quantification of bacteria using 16S ribosomal sequences have significantly advanced our understanding of this system (2). The application of quantitative PCR (qPCR) to 16S rRNA technology has been validated for the quantification of bacterial groups and specific bacterial species within complex bacterial populations (5, 20, 32).
In conventionally reared mice, the majority of enteric pathogens are unable to persistently colonize the gastrointestinal (GI) tract, possibly due to the presence of an intact microbiota. However, Salmonella enterica serovar Typhimurium is capable of both colonization and infection of the murine small intestine. Salmonella enterica is a gram-negative pathogen that can infect diverse mammalian hosts. In humans, Salmonella enterica serovar Typhimurium is a common cause of food-borne illness and causes gastroenteritis, while in mice it predominantly causes a systemic “typhoid-like” illness. Here we show that virulent S. enterica serovar Typhimurium persistently colonizes FvB mice and causes enteritis, although other mouse models suggest the requirement for antibiotic pretreatment to achieve this outcome (4). We hypothesized that S. enterica serovar Typhimurium infection of the mouse gut alters the commensal microbiota, either directly through Salmonella-commensal interaction or indirectly through the host mucosal immune response to this pathogen. This study investigates this hypothesis and finds that wild-type S. enterica serovar Typhimurium infection interferes with host “colonization resistance,” as reflected by persistent Salmonella colonization. Salmonella infection also disrupts the normal composition of the gut microbiota, and this disruption is associated with Salmonella virulence factors that have been shown to induce inflammatory mucosal host responses.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Salmonella enterica serotype Typhimurium strain 14028 (ATCC) was cultured aerobically at 37°C in Luria-Bertani (LB) broth as described previously (28). Briefly, S. enterica serovar Typhimurium was grown in a 10-ml standing culture for 3.5 h at 37°C to select for the most motile bacteria. The top 5 ml of culture was transferred to 45 ml of prewarmed LB broth and grown with shaking for 1.5 h at 37°C. The quantity of S. enterica serovar Typhimurium was determined using a Petroff-Hausser chamber (Hausser Scientific, PA), and the appropriate dose was prepared in sterile 0.2 M phosphate buffer (pH 8). Isogenic Salmonella pathogenicity island 1 (SPI1) and SPI2 mutants (TK93 and 5-SAT, respectively) were obtained from Samuel Miller (University of Washington, Seattle, WA) (15, 21).
Animal experiments.
Male and female wild-type FvB mice were obtained from Taconic Laboratories and used to generate litters for these experiments. Animals were bred and housed under specific-pathogen-free conditions in the Medical College of Wisconsin Biomedical Resource Center vivarium. All animal-related experiments and procedures were approved by the animal care and use committee at the Medical College of Wisconsin. In order to control for maternal effects on intestinal colonization, each experiment used animals from a single litter. Successive litters from individual breeding pairs were used for each time course. To control for environmental effects on the intestinal microbiota, litters were housed together from birth until 5 weeks of age. At 5 weeks of age, animals were deprived of food overnight. Half of the animals in each litter were inoculated with S. enterica serovar Typhimurium by intragastric gavage, while the other half were given vehicle alone. Animals were given either 107 (the 50% lethal dose [LD50]) (28) or 108 (10× LD50) CFU S. enterica serovar Typhimurium, as indicated. Surviving animals were sacrificed after 1, 3, 7, or 30 days, as indicated. The terminal 1.5 cm of ileum was fixed in Carnoy's solution (Fisher Scientific) for histology and fluorescence in situ hybridization (FISH) studies. The intestinal tract was removed and separated into the distal small intestine (DSI; the distal 15 cm), cecum, and large intestine (LI). Samples were used for qPCR. Experiments involving SPI1 and SPI2 mutants were performed as described above, using inocula of 108 CFU Salmonella. Animals were sacrificed after 3 days.
FISH.
Mouse terminal ileum was fixed in Carnoy's fixative and processed as described previously (8). Three-micrometer sections were mounted on slides, and FISH was performed as described previously (8), using a combination of a 6-carboxyfluorescein (FAM)-labeled oligonucleotide probe for Salmonella enterica (Sal998 [FAM-TCTCTGGATTCTTCTGTGGA]) along with a Texas Red (TR)-labeled universal bacterial probe (Bact338 [TR-GCTGCCTCCGTAGGAGT]) (Operon Technologies, Huntsville, AL). Briefly, slides were deparaffinized, dried, and hybridized with the indicated probe combinations for 90 min at 50°C in hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl, pH 7.4, 0.05% sodium dodecyl sulfate). Slides were washed for 5 min at 50°C in wash buffer (0.9 M NaCl, 20 mM Tris-HCl, pH 7.4, 0.01% sodium dodecyl sulfate), rinsed in water, and allowed to air dry. Tissue sections were mounted with coverslips, using Vectashield mounting medium (Vector Laboratories, Burlingame, CA) for fluorescence. Slides were viewed by fluorescence microscopy using a Nikon E400 upright microscope. Images were captured using a Photometrics CoolSnap ES charge-coupled device camera and analyzed using Metaview software (Universal Imaging Corporation, Molecular Devices).
Histology.
Three-micrometer sections of Carnoy's fixative-fixed terminal ileum were mounted on slides and stained with hematoxylin and eosin. Slides were examined by an anatomic pathologist, using a Nikon E400 upright microscope. Images were captured using a Spot camera and analyzed using Spot software, version 3.5.4 (Diagnostic Instruments, Inc.).
Bacterial genomic DNA extraction.
The DSI, ceca, and LI isolated from the Salmonella infection experiments were weighed and then homogenized using a Polytron PT 10-35 homogenizer (Kinematica Switzerland) in 2 ml sterile phosphate-buffered saline. Bacterial genomic DNA was extracted from the DSI, cecum, and LI by use of a Qiagen stool kit (Qiagen) according to the kit directions, using the optional high-temperature step.
Quantitative real-time PCR amplification of 16S rRNA gene sequences.
The abundance of specific intestinal bacterial groups was measured by qPCR using a MyiQ single-color real-time PCR detection system (Bio-Rad, Hercules, CA) with group-specific 16S rRNA gene primers (Operon Technologies, Huntsville, AL) (Table 1). A short segment of the 16S rRNA gene (200 to 300 bp) was specifically amplified by real-time PCR, using the conserved 16S rRNA-specific primer pair UniF340 and UniR514 (Table 1), to determine the total amount of commensal bacteria in each intestinal segment. The real-time PCR program started with an initial step at 95°C for 3 min, followed by 40 cycles of 10 s at 95°C and 45 s at 63°C (Table 1). Data were acquired in the final step at 63°C. The real-time PCRs were done using IQ SYBR green supermix (Bio-Rad). Using the same genomic DNA from each sample, real-time PCRs were completed using group-specific primers to determine the amount of bacteria in each of the following major groups: Eubacterium rectale/Clostridium coccoides, Lactobacillus sp., Bacteroides sp., mouse intestinal Bacteroides, segmented filamentous bacteria, Enterobacteriaceae, S. enterica serovar Typhimurium, Clostridium perfringens, and Helicobacter (Table 1). Bacterial numbers were determined using standard curves constructed with reference bacteria specific for each bacterial group analyzed (Table 1). qPCR measures the number of 16S rRNA gene copies per sample, not actual bacterial numbers or CFU. Nevertheless, these values are directly related and correlate well. To validate qPCR for the quantification of Salmonella from a mixed bacterial population, Salmonella was quantified by both culture and qPCR after growth in a mixed Salmonella-Escherichia coli culture. LB broth was inoculated with either S. enterica serovar Typhimurium or E. coli or coinoculated with both S. enterica serovar Typhimurium and E. coli and then grown at 37°C, with shaking, overnight. Serial dilutions of the cultures were plated on LB (to quantify total CFU) and SS (to quantify total Salmonella) agar. Bacterial genomic DNA was isolated from the cultures, and Salmonella was quantified by qPCR as described above.
TABLE 1.
16S rRNA gene group-specific and kingdom-specific primers for qPCRa
| Group | Reference strain | Primer | Sequence (5′ to 3′) | Temp for last step (°C) | Reference |
|---|---|---|---|---|---|
| Eubacteria (all bacteria) | Ruminococcus productus | UniF340 | ACTCCTACGGGAGGCAGCAGT | 63 | 1 |
| ATCC 27340D | UniR514 | ATTACCGCGGCTGCTGGC | 63 | ||
| Eubacterium rectale/ | Ruminococcus productus | UniF338 | ACTCCTACGGGAGGCAGC | 60 | 12 |
| Clostridium coccoides | ATCC 27340D | C.cocR491 | GCTTCTTAGTCAGGTACCGTCAT | 60 | |
| Lactobacillus/Lactococcus | Lactobacillus acidophilus | LabF362 | AGCAGTAGGGAATCTTCCA | 56 | 27 |
| ATCC 4357D | LabR677 | CACCGCTACACATGGAG | 56 | ||
| Bacteroides | Bacteroides fragilis | BactF285 | GGTTCTGAGAGGAGGTCCC | 61 | 10 |
| ATCC 25285D | UniR338 | GCTGCCTCCCGTAGGAGT | 61 | ||
| Mouse intestinal Bacteroides | MIB plasmid CT11-6 | Uni516F | CCAGCAGCCGCGGTAATA | 58 | 29 |
| MIBR677 | CGCATTCCGCATACTTCTC | 58 | |||
| Segmented filamentous bacteria | SFB plasmid CTL5-6 | SFB736F | GACGCTGAGGCATGAGAGCAT | 58 | 30 |
| SFB844R | GACGGCACGGATTGTTATTCA | 58 | |||
| Salmonella enterica serovar | ATCC 700720-D | Sal454 | TGTTGTGGTTAATAACCGCA | 56 | 17 |
| Typhimurium | Uni785R | GACTACCAGGGTATCTAATCC | 56 | ||
| Enterobacteriaceae | Escherichia coli ATCC | Uni515F | GTGCCAGCMGCCGCGGTAA | 67 | 16 |
| 10798D | Ent826R | GCCTCAAGGGCACAACCTCCAAG | 67 | ||
| Clostridium perfringens | Clostridium perfringens | Cperf165F | CGCATAACGTTGAAAGATGG | 61 | 34 |
| ATCC 13124D-5 | Cperf269R | CCTTGGTAGGCCGTTACCC | 61 | ||
| Helicobacter | Helicobacter pylori | Hp 547F | CTTAACCATAGAACTGCATTTGAAACTAC | 57 | 32 |
| ATCC 700392D-5 | Hp 665R | GGTCGCCTTCGCAATGAGTA | 57 |
Primer sets for qPCR were designed based on published probes specific for the different bacterial groups in combination with a universal primer. Groups were defined in ARB software (18) based on the published probe in combination with a suitable universal primer or a pair of published primers. The compatibility of primer combinations for qPCR was tested by Primer Designer 5 (Sci Ed Programs). The primer sets were tested for sensitivity and specificity against a wide panel of bacterial genomic DNAs and showed minimal cross-reactivity.
Statistical analysis.
Analyses were performed using the PROC MIXED feature of SAS 9.1.3 (SAS Institute, Cary, NC). A separate analysis was performed for each dose/time point/gut segment combination. Each of these combinations was considered to be a separate experiment. A mixed model was fitted to the data to account for the repeated-measures design. The treatment, the bacterial group, and their interaction were fixed effects, while the animal was a random effect. To determine whether log counts of any bacterial groups were different between infected and control animals, an F test of the join effect of treatment and bacterial group was done. If the null hypothesis was rejected for this question, an F test of treatment-bacterial group interaction was done to determine whether the changes were uniform across bacterial groups. If the null hypothesis was rejected for this question, the question of which bacterial groups were changing was analyzed. The treatment effect within each bacterial group, with corresponding standard errors and t test results, was estimated from the model.
RESULTS
Peroral S. enterica serovar Typhimurium infection results in ongoing colonization of the mouse intestinal tract.
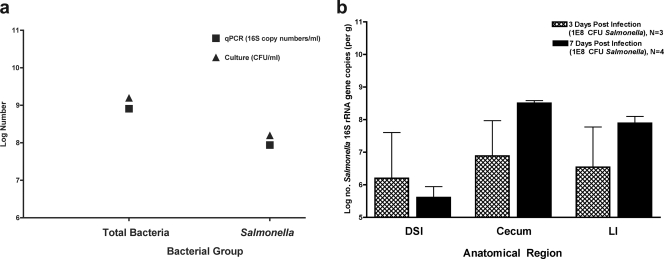
While most bacteria delivered orally to mice cannot colonize the murine intestinal tract, Salmonella persists for as long as 30 days postinoculation. Consistent with the approach used to quantify members of the indigenous intestinal microbiota, qPCR was used to determine and quantify the presence of Salmonella in the intestinal tract. To validate the use of qPCR for quantification of Salmonella in a mixed bacterial population, S. enterica serovar Typhimurium was cocultured with E. coli in LB broth overnight. Total bacteria and total Salmonella were quantified by both selective culture and qPCR. Although qPCR measures the number of 16S rRNA gene copies per sample, not actual bacterial numbers or CFU, these values are directly related and correlate well (Fig. 1a). After oral inoculation with 108 CFU (high dose; 10× LD50) of wild-type S. enterica serovar Typhimurium, Salmonella persisted in each segment of the lower GI tract for up to 7 days postinoculation (Fig. 1b). High-dose peroral Salmonella infection was lethal to mice within 7 to 10 days. To investigate longer-term colonization, mice were inoculated with a lower dose of Salmonella (107 CFU/mouse [the LD50]). After inoculation with the lower dose, Salmonella was detected as long as 30 days postinfection in the DSI (see Fig. 8a). The continued presence of Salmonella in the intestinal lumen appears to be due to persistent colonization. While it is possible that the presence of intestinal Salmonella results from systemic spread and reinfection from the bloodstream, we do not find evidence for this in this experimental system. For example, analysis of the intestinal microbiota after systemic (intraperitoneal) infection with S. enterica serovar Typhimurium does not show the presence of this pathogen in the gut or evidence of mucosal inflammation in the Peyer's patches (not shown). The ability of Salmonella to persistently colonize the intestinal tracts of FvB mice without antibiotic pretreatment suggests that S. enterica serovar Typhimurium infection of FvB mice is a useful model for the study of infectious colitis and its impact on the indigenous microbiota.
FIG. 1.
Validation of qPCR for quantification of Salmonella and persistent colonization of the murine intestinal tract by Salmonella. (a) LB broth was coinoculated with S. enterica serovar Typhimurium and E. coli, which were grown at 37°C. An aliquot of culture was dilution plated on LB (to enumerate total bacteria) or SS (to enumerate Salmonella) agar. Total bacterial genomic DNA was isolated from an aliquot of culture and analyzed by qPCR to enumerate total bacteria and Salmonella. Black squares denote log numbers of bacteria determined by qPCR. Black triangles denote log numbers of bacteria determined by selective culture. (b) Mice were orally inoculated with either 108 CFU S. enterica serovar Typhimurium or buffer alone and then sacrificed at 3 or 7 days postinoculation. The intestinal tract was removed and divided. Bacterial genomic DNA was isolated from the DSI, cecum, and LI and analyzed by qPCR for quantification of Salmonella. No Salmonella cells were seen in control mice. Hatched bars represent infected mice at 3 days postinoculation. Black bars represent infected mice at 7 days postinoculation.
FIG. 8.
Quantitative analysis of the intestinal microbiota after intestinal clearance of S. enterica serovar Typhimurium. Mice were inoculated with 107 CFU S. enterica serovar Typhimurium and sacrificed after 30 days. The intestinal tract was removed and divided into the DSI, cecum, and LI. Bacterial genomic DNA was isolated from each segment, and qPCR analysis was performed to determine the abundance of specific commensal bacterial groups in the DSI (a) and cecum (b). White bars represent uninfected controls. Black bars represent Salmonella-infected mice. Salmonella infection did not appear to affect the bacterial numbers in any segment of the gut (P > 0.05). BT, below the detection threshold of qPCR. Erec, Eubacterium rectale/Clostridium coccoides; Lact, Lactobacillus sp.; Bact, Bacteroides sp.; MIB, mouse intestinal Bacteroides; Sfb, segmented filamentous bacteria; Ent, Enterobacteriaceae; C. perf, Clostridium perfringens; Salm, S. enterica serovar Typhimurium.
High-dose peroral S. enterica serovar Typhimurium infection causes enteritis and diarrhea.
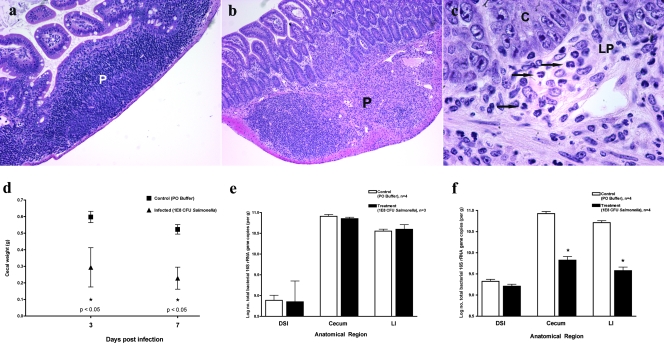
Although they were not pretreated with streptomycin or other antibiotics, Salmonella-infected mice presented evidence of infectious enteritis similar to that described for antibiotic pretreatment models (4), although the onset of enteritis is accelerated in the antibiotic pretreatment models. Three days after peroral inoculation with S. enterica serovar Typhimurium, there was early evidence of infection, with infiltration of the Peyer's patches by neutrophils (not shown). By 7 days postinfection, a mixed cellular infiltrate of mononuclear cells and neutrophils infiltrated the Peyer's patches (Fig. 2b) and the lamina propria (Fig. 2c [high magnification]). A normal Peyer's patch from an uninfected mouse terminal ileum is shown for comparison (Fig. 2a). By 3 days postinfection, the ceca of these mice had already appreciably decreased in weight, and by day 7 they appeared shrunken and weighed significantly less than those of uninfected controls (Fig. 2d). Diarrhea was noted at 7 days of infection, as evidenced by the LI being filled with liquid contents but no formed stool, staining of the perianal and tail regions with liquid fecal material, and accumulation of loose and liquid fecal material in the cage bedding (not shown). qPCR measurements of total bacteria paralleled these observations, showing a loss of 95% of the total quantity of commensal bacteria in the cecum and LI (Fig. 2f) at 7 days postinoculation. Changes in cecal weight did not appear to be linked directly to loss of bacterial numbers, since the weight decreases appeared at 3 days postinfection but the total number of bacteria was unchanged at this time point (Fig. 2e). These findings demonstrate the development of gross and microscopic enteritis in these mice over a period of 1 week.
FIG. 2.
Characterization of enteric salmonellosis. Mice were orally inoculated with 108 CFU S. enterica serovar Typhimurium or buffer alone and then sacrificed after either 3 or 7 days. The mouse terminal ileum was removed and fixed in Carnoy's fixative. Hematoxylin and eosin staining was performed on terminal ileum sections from control (a) and Salmonella-infected (b and c) mice at 7 days postinfection. P, Peyer's patch; C, crypt epithelium; LP, lamina propria. Arrows indicate the locations of neutrophils. (d) The ceca of control and infected mice were removed and weighed for comparison at both 3 and 7 days postinfection. Black squares represent uninfected control mice. Black triangles represent Salmonella-infected mice. Bacterial genomic DNA was isolated from the DSI, ceca, and LI of control and infected mice at 3 (e) and 7 (f) days postinoculation and analyzed by qPCR for total bacteria. White bars represent uninfected controls. Black bars represent Salmonella-infected mice. *, P < 0.05 (Student's t test).
High-dose peroral S. enterica serovar Typhimurium infection alters the intestinal microbiota.
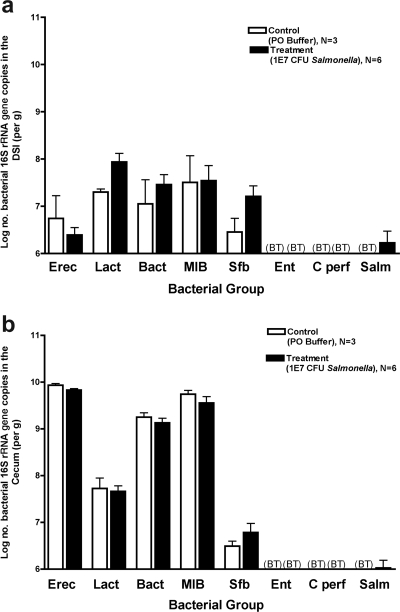
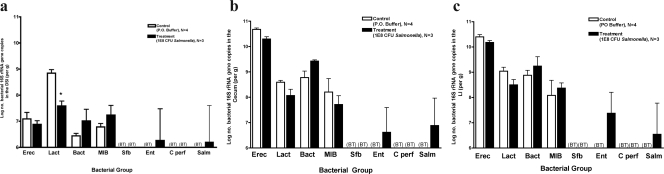
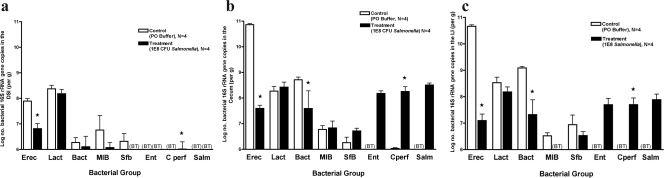
To further characterize the impact of S. enterica serovar Typhimurium infection on the intestinal microbiota, quantitative analysis of several of the most common bacterial groups comprising the microbiota was done. At 3 days post-Salmonella inoculation, notable changes in the microbiota were present primarily in the DSI, with significant decreases in the Lactobacillus/Enterococcus group (Fig. 3a). However, after 1 week, profound decreases in the Eubacterium rectale/Clostridium coccoides group were evident throughout all sections of the intestine (Fig. 4a, b, and c), along with corresponding increases in the C. perfringens group. The cecum and LI demonstrated decreases in Bacteroides, but this bacterial group was stable in the DSI. Dramatic changes in the most abundant bacterial group, the Eubacterium rectale/Clostridium coccoides group, were evident at 1 week, suggesting that this group may be most susceptible during the diarrheal host response.
FIG. 3.
Quantitative analysis of intestinal microbiota 3 days after infection with 108 CFU S. enterica serovar Typhimurium. Mice were inoculated with 108 S. enterica serovar Typhimurium organisms and sacrificed after 3 days. The intestinal tract was removed and divided into the DSI, cecum, and LI. Bacterial genomic DNA was isolated from each segment, and qPCR analysis measured the abundance of specific commensal bacterial groups in the DSI (a), cecum (b), and LI (c). White bars represent uninfected controls. Black bars represent Salmonella-infected mice. In the DSI, Salmonella infection did affect bacterial counts (P < 0.05), and the effect was not uniform across groups (P < 0.05). The asterisk represents the post hoc t test for the Lactobacillus sp. group (P < 0.005). Salmonella infection did not appear to affect the cecum or LI (P > 0.05). BT, below the detection threshold of qPCR. Erec, Eubacterium rectale/Clostridium coccoides; Lact, Lactobacillus sp.; Bact, Bacteroides sp.; MIB, mouse intestinal Bacteroides; Sfb, segmented filamentous bacteria; Ent, Enterobacteriaceae; C. perf, Clostridium perfringens; Salm, S. enterica serovar Typhimurium.
FIG. 4.
Quantitative analysis of intestinal microbiota 7 days after infection with 108 CFU S. enterica serovar Typhimurium. Mice were inoculated with 108 S. enterica serovar Typhimurium organisms and sacrificed after 7 days. The intestinal tract was removed and divided into the DSI, cecum, and LI. Bacterial genomic DNA was isolated from each segment, and qPCR analysis measured the abundance of specific commensal bacterial groups in the DSI (a), cecum (b), and LI (c). White bars represent uninfected controls. Black bars represent Salmonella-infected mice. Salmonella infection did affect bacterial counts in the DSI (P < 0.0001), cecum (P < 0.0001), and LI (P < 0.0001), and the effects were not uniform across groups (P < 0.0001 for all segments). Asterisks represent the post hoc t test for the designated groups (P < 0.05). BT, below the detection threshold of qPCR. Erec, Eubacterium rectale/Clostridium coccoides; Lact, Lactobacillus sp.; Bact, Bacteroides sp.; MIB, mouse intestinal Bacteroides; Sfb, segmented filamentous bacteria; Ent, Enterobacteriaceae; C. perf, Clostridium perfringens; Salm, S. enterica serovar Typhimurium.
Salmonella organisms are present but do not supersede the indigenous microbiota of the intestinal tract.
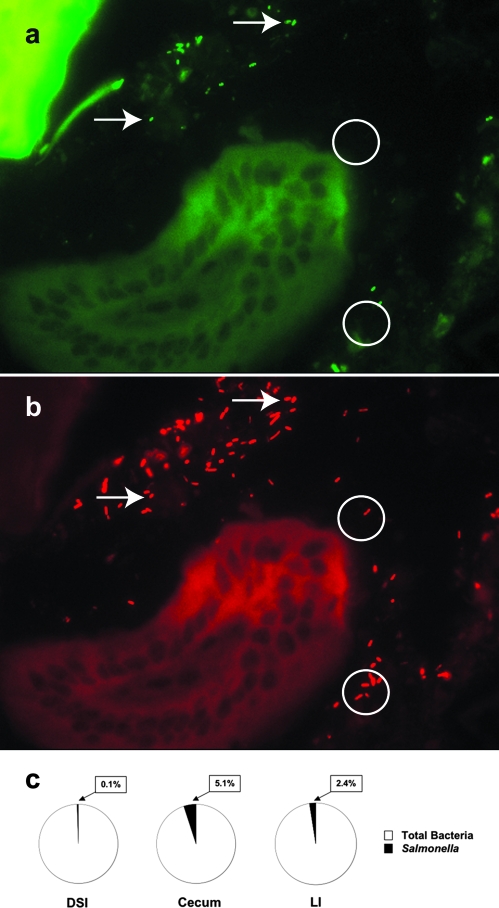
Seven days after infection with 108 S. enterica serovar Typhimurium organisms, the terminal ilea of mice were analyzed for the presence of Salmonella by FISH (Fig. 5). Salmonella cells were identified by hybridization with a fluorescein isothiocyanate-labeled oligonucleotide probe generated from a Salmonella-specific 16S rRNA sequence (Fig. 5a). Total bacteria were identified by hybridization with a TR-labeled oligonucleotide common to 16S rRNA bacterial sequences (Fig. 5b). Consistent with the quantitative findings, Salmonella cells were present within the gut lumen (Fig. 5a), but not in high numbers, and were found in much lower abundance than the indigenous microbiota (Fig. 5b and c). qPCR to quantify the total microbiota and the abundance of Salmonella in the DSI, cecum, and LI 7 days after high-dose infection was consistent with the FISH finding (Fig. 5c). While Salmonella was present, it had not replaced or superseded the indigenous microbiota.
FIG. 5.
Localization of Salmonella in the terminal ileum. The mouse terminal ileum was removed from mice at 7 days post-peroral inoculation with 108 CFU S. enterica serovar Typhimurium and fixed in Carnoy's fixative. Three-micrometer sections were cut and analyzed by FISH to localize bacteria within the tissue section. Sections of small intestinal tissue were cohybridized with a combination of FAM-Sal and TR-Bact338 probes, which enabled visualization of the distribution of both Salmonella (a) and the total indigenous microbiota (b) within the small intestinal lumen. FAM-Sal specifically hybridizes to Salmonella (a), while TR-Bact338 shows total bacteria (b). Arrows show examples of Salmonella in each field. Circles illustrate examples of non-Salmonella indigenous bacteria. (c) qPCR was used to quantify total bacteria and Salmonella from the DSI, ceca, and LI of infected mice. The black section of each pie chart represents the percentage of the total microbiota comprised by Salmonella.
Impact of SPI1 and SPI2 mutants on the indigenous intestinal microbiota.
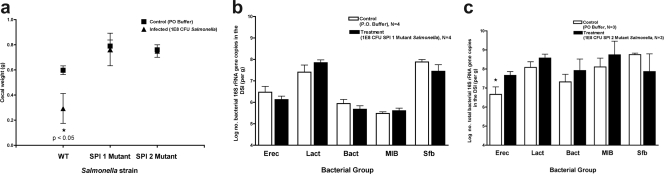
To begin to investigate the specific mechanisms by which Salmonella infection alters the microbiota, specific Salmonella type 3 secretion system mutants were used for peroral infections. SPI1 mutants have wild-type virulence when given systemically but are attenuated when given orally (6). They are unable to directly invade intestinal epithelial cells but may be taken up by intestinal dendritic cells (26). Oral infection with SPI1 mutants does not result in local mucosal inflammation (data not shown). Three days after oral inoculation with 108 CFU of TK93, a SPI1 mutant, the mouse intestinal tract was examined for evidence of inflammation, diarrhea, and any changes in the composition of the indigenous microbiota. Grossly, the intestinal tract looked normal, with no change in cecal weight (Fig. 6a). Histologic examination of the terminal ileum did not demonstrate evidence of local inflammation (not shown). Analysis of the microbiota revealed no changes in response to TK93 inoculation (Fig. 6b). In addition, there was no evidence of mutant Salmonella remaining in any section of the intestinal tract (not shown). Similar experiments were done with 5SAT, a SPI2 mutant. SPI2 mutants should be able to invade the intestinal mucosa but unable to survive within the phagolysosome (13). Three days after oral inoculation with 108 CFU of 5SAT, we found neither changes in cecal weight (Fig. 6a) nor histologic evidence of inflammation (not shown). However, the mutant bacteria could still be found in the LI, and statistically significant changes in the indigenous microbiota were noted in the DSI (Fig. 6c).
FIG. 6.
Impact of SPI1 and SPI2 Salmonella mutants on the intestinal microbiota. Mice were inoculated perorally with 108 CFU of S. enterica serovar Typhimurium TK93 (SPI1 mutant) or 5SAT (SPI2 mutant) and were sacrificed after 3 days. The intestinal tract was removed and divided into the DSI, cecum, and LI. (a) Cecal weights were obtained and compared between control and infected mice. Black squares represent uninfected control mice. Black triangles represent Salmonella-infected mice. (b) Bacterial genomic DNA was isolated from the DSI of mice infected with the SPI1 mutant (b) and the SPI2 mutant (c), and qPCR analysis determining the abundance of specific commensal groups was performed. White bars represent uninfected controls. Black bars represent Salmonella-infected mice. The asterisk represents the post hoc t test for the designated group (P < 0.05). Erec, Eubacterium rectale/Clostridium coccoides; Lact, Lactobacillus sp.; Bact, Bacteroides sp.; MIB, mouse intestinal Bacteroides; Sfb, segmented filamentous bacteria.
Low-dose peroral S. enterica serovar Typhimurium infection alters the intestinal microbiota without diarrhea.
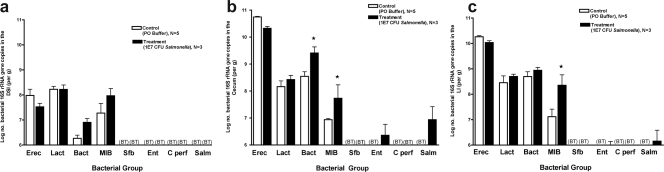
High-dose oral S. enterica serovar Typhimurium inoculation was lethal to mice, not allowing investigation of long-term colonization and its impact on the microbiota. To investigate whether S. enterica serovar Typhimurium is capable of inducing and maintaining changes in the microbiota during long-term colonization in the absence of severe enteritis or diarrhea, mice were challenged with a lower dose (107 CFU/mouse [the LD50]) of S. enterica serovar Typhimurium, and surviving mice were analyzed at 1, 7, and 30 days postinfection. There was no evidence of diarrhea throughout this time course, and cecal weights did not differ between infected and control mice (not shown). While examination of the composition of the microbiota did not reveal significant differences at 1 day postinoculation (not shown), changes were evident 1 week after infection (Fig. 7b and c). There were significant increases in both Bacteroides groups measured, i.e., total Bacteroides and mouse intestinal Bacteroides. It is clear that a lower inoculum of S. enterica serovar Typhimurium results in alterations of the microbiota, even in the absence of overt enteritis. However, if the inoculum is reduced further, to 106 CFU per mouse, there are no changes in the indigenous microbiota, and Salmonella is cleared from the GI tract (not shown).
FIG. 7.
Quantitative analysis of the intestinal microbiota after 7 days of infection with 107 CFU S. enterica serovar Typhimurium. Mice were inoculated perorally with 107 CFU S. enterica serovar Typhimurium and sacrificed after 7 days. The intestinal tract was removed and divided into the DSI, cecum, and LI. Bacterial genomic DNA was isolated from each segment, and qPCR analysis was performed to determine the abundance of specific commensal bacterial groups in the DSI (a), cecum (b), and LI (c). White bars represent uninfected controls. Black bars represent Salmonella-infected mice. Salmonella infection did affect bacterial counts in the cecum (P < 0.005) and the LI (P < 0.05), and the effects were not uniform across groups. Asterisks represent the post hoc t test for the designated groups (P < 0.005). Salmonella infection did not appear to affect the bacterial counts in the small intestine (P > 0.05). BT, below the detection threshold of qPCR. Erec, Eubacterium rectale/Clostridium coccoides; Lact, Lactobacillus sp.; Bact, Bacteroides sp.; MIB, mouse intestinal Bacteroides; Sfb, segmented filamentous bacteria; Ent, Enterobacteriaceae; C. perf, Clostridium perfringens; Salm, S. enterica serovar Typhimurium.
Clearance of S. enterica serovar Typhimurium infection results in reversion of the microbiota to that of uninfected mice.
Thirty days after inoculation, infected mice did not show the presence of Salmonella in their feces (not shown). The ceca (Fig. 8b) and LI (not shown) of these mice did not show evidence of Salmonella colonization. A comparison between the microbiotas of the infected mice and uninfected controls did not show significant differences at these sites. This reversion of the indigenous microbiota to baseline demonstrates that bacterial enteritis does not permanently alter the microbiota. Complete clearance of the pathogen did not appear to be required for this reversion, as demonstrated by the DSI. Colonization by Salmonella persisted in the small intestine at 30 days postinoculation, although at a low abundance (Fig. 8a). Comparisons between infected and control mice demonstrated no significant differences in the compositions of the small intestinal microbiota.
DISCUSSION
The maintenance of the microbial ecology of the gut involves a complex web of bacterium-host and bacterium-bacterium interactions. While much is known about bacterial interactions with the host at mucosal surfaces for specific pathogens, including S. enterica serovar Typhimurium, the effect of enteric infection on the microbial ecology of the intestine is not understood. In this paper, we investigated the effect of enteric S. enterica serovar Typhimurium infection on the murine intestinal microbiota. Using a murine Salmonella enteritis model that did not depend on antibiotic pretreatment, we investigated the impact of infectious enteritis on the intestinal microbiota and found that Salmonella infection caused acute enteritis, diarrhea, and profound changes in the intestinal microbial ecology. Changes in the microbiota were associated with the mucosal host response (diarrhea) and appeared to require both SPI1 and SPI2 type 3 secretion systems.
The protective capacity of the gut microbiota in mice is well documented. Very few exogenously delivered bacteria, either commensal or pathogen, are able to persistently colonize the murine intestinal tract. Even in the case of pathogens that cause systemic infection gaining access through the gut, these pathogens do not successfully persist in the gut lumen. Altering the biota by antibiotics renders the host much more susceptible to enteric infection, characterized by greater translocation of pathogens, faster systemic spread, and lower infectious doses. S. enterica serovar Typhimurium is an exception to some of these generalizations. In FvB mice, S. enterica serovar Typhimurium is able to overcome colonization resistance without antibiotic pretreatment, resulting in persistent colonization and localized enteritis.
The severe enteritis noted after high-dose S. enterica serovar Typhimurium infection was accompanied by diarrhea and accompanying losses of 95% of the total indigenous bacteria from the cecum and LI as well as by profound changes in the microbiota composition. Notably, these changes were associated with the presence of relatively small numbers of Salmonella in the gut lumen, so it appears that the pathogen does not need to outcompete the indigenous biota to exert its effects. Alterations in the biota may be ascribed to host responses to the pathogen rather than to the presence of the pathogen itself. These profound changes were likely due to the diarrheal host response, particularly the dramatic loss of the Eubacterium rectale/Clostridium coccoides group, which appeared to be selective in susceptibility during this time frame of the innate host response. Changes in the biota could not be ascribed entirely to diarrhea, since some changes were evident at 3 days postinfection, prior to the gross evidence of diarrhea. Similarly, disruption of the microbiota composition was evident several days after low-dose inoculation, and neither diarrhea nor a decrease in cecal size or weight was seen during the low-dose experiment. Together, these data suggested that additional pathogen-host and/or pathogen-commensal interactions are involved in shifting the composition of the biota and that both host response and microbiota alteration are multiphasic and possibly dose dependent.
Additional evidence suggests the importance of the mucosal host response. First, the presence of Salmonella alone, without evidence of ongoing inflammation, as seen in the DSI at 30 days postinfection, did not impact the biota. In these mice, the microbiota largely reverted to that of uninfected controls. Evidence from the use of Salmonella strains with mutations in specific virulence factors demonstrates the importance of the pathogen-host interaction in disrupting intestinal microbial ecology. S. enterica serovar Typhimurium lacking the SPI1 type 3 secretion system, while able to gain access to the systemic circulation via the gut, was unable to generate a local mucosal immune response and unable to persist in the gut. Peroral infection with this mutant did not result in alterations in the microbiota, induction of local inflammation, or persistence in the intestine. Infection with a SPI2 mutant gave mixed results. SPI2 mutants should be able to invade through the intestinal mucosa, inducing local immune responses. We anticipated that this mutant might be able to attract neutrophils to the Peyer's patches, to persistently colonize the intestinal tract, and to alter the intestinal microbiota. We did not see evidence of neutrophilic infiltration or changes in cecal weight with this mutant and saw only a minimal ability to persist in the intestine. However, we did note some disruption of the indigenous microbiota that was not seen with the SPI1 mutant. This suggests that the SPI1 type 3 secretion system is essential to Salmonella's ability to disrupt the microbiota but not sufficient for the cumulative profound effects that are seen with the wild-type organism. It has been reported that SPI2 mutants may demonstrate defective expression of SPI1 (9), so we cannot rule out the possibility that the incomplete effect of this mutant could be associated with a similar problem. Recently published work also highlights the importance of the host immune response in the ability of pathogens to alter the intestinal microbiota. Stecher et al. (31) have shown that after antibiotic treatment, S. enterica serovar Typhimurium is able to colonize the GI tract and prevent recovery of the indigenous microbiota, secondary to induction of local host immune responses. Additional evidence from Lupp et al. (19), who used a Citrobacter rodentium infection model, has similarly demonstrated the importance of host inflammation in the disruption of intestinal microbial ecology.
After clearance of Salmonella from the cecum and LI at 30 days postinfection, the microbiota has also reverted to that of uninfected controls. The reversion of the intestinal microbial ecology to that of uninfected controls is intriguing, demonstrating that disruptions in the composition of the microbiota by infectious colitis are temporary and that the host gravitates back to a set point, which is likely influenced by a combination of genetic, immune, nutritional, and environmental factors of the host. This may explain the difficulty seen in attempts to effect long-term changes in the intestinal microbiota by the use of probiotics, which have not been shown to persist in the gut after discontinuance of administration of the bacteria.
What is the significance of the alterations in the microbiota triggered by Salmonella enteritis? It is likely, in part, that changes in the biota represent “collateral damage” secondary to the host immune response attempting to eliminate an invading pathogen. Effective enteric pathogens, such as S. enterica serovar Typhimurium, may take advantage of this response. In fact, this may be considered a mechanism of effective pathogenesis. The host, in generating an immune response to eliminate a pathogen, may inadvertently damage its protective microbial ecosystem. Although most pathogens may be cleared by this host response, pathogens that are resistant to this mechanism of host defense may trigger this response to gain a foothold and colonize the intestinal tract.
Acknowledgments
We thank Aniko Szabo, Division of Biostatistics, Medical College of Wisconsin, for statistical analysis of the data. We thank Samuel Miller for the Salmonella mutants. We thank Charles Bevins and Joseph Barbieri for critically reading the manuscript.
Microscope slides were examined by N.H.S.
This work was supported by Public Health Service grant AI057757 (N.H.S.) from the National Institutes of Health and by the Diabetes Foundation Netherlands (N.A.B.).
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 26 December 2007.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ Microbiol. 561919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 3071915-1920. [DOI] [PubMed] [Google Scholar]
- 3.Baker, G. C., J. J. Smith, and D. A. Cowan. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55541-555. [DOI] [PubMed] [Google Scholar]
- 4.Barthel, M., S. Hapfelmeier, L. Quintanilla-Martinez, M. Kremer, M. Rohde, M. Hogardt, K. Pfeffer, H. Russmann, and W. D. Hardt. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 712839-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartosch, S., A. Fite, G. T. Macfarlane, and M. E. McMurdo. 2004. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effect of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 703575-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumler, A. J., R. M. Tsolis, P. J. Valentine, T. A. Ficht, and F. Heffron. 1997. Synergistic effect of mutations in invA and lpfC on the ability of Salmonella typhimurium to cause murine typhoid. Infect. Immun. 652254-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg, R. D. 1981. Promotion of the translocation of enteric bacteria from the gastrointestinal tracts of mice by oral treatment with penicillin, clindamycin, or metronidazole. Infect. Immun. 33854-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canny, G., A. Swidsinski, and B. A. McCormick. 2006. Interactions of intestinal epithelial cells with bacteria and immune cells: methods to characterize microflora and functional consequences. Methods Mol. Biol. 34117-35. [DOI] [PubMed] [Google Scholar]
- 9.Coombes, B. K., M. E. Wickham, M. J. Lowden, N. F. Brown, and B. B. Finlay. 2005. Negative regulation of Salmonella pathogenicity island 2 is required for contextual control of virulence during typhoid. Proc. Natl. Acad. Sci. USA 10217460-17465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dore, J., A. Sghir, G. Hannequart-Gramet, G. Corthier, and P. Pochart. 1998. Design and evaluation of a 16S rRNA-targeted oligonucleotide probe for specific detection and quantitation of human faecal Bacteroides populations. Syst. Appl. Microbiol. 2165-71. [DOI] [PubMed] [Google Scholar]
- 11.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 3081635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 643336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Ganks, A. Vazques-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30163-174. [DOI] [PubMed] [Google Scholar]
- 14.Hooper, L. V., T. Midtvedt, and J. I. Gordon. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22283-307. [DOI] [PubMed] [Google Scholar]
- 15.Kimbrough, T. G., and S. I. Miller. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. USA 9711008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lane, D. J. 1991. rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY.
- 17.Lin, C. K., and H. Y. Tsen. 1996. Use of two 16S DNA targeted oligonucleotides as PCR primers for the specific detection of Salmonella in foods. J. Appl. Bacteriol. 80659-666. [DOI] [PubMed] [Google Scholar]
- 18.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 321363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupp, C., M. L. Robertson, M. E. Wickham, I. Sekirov, O. L. Champion, E. C. Gaynor, and B. B. Finlay. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2119-129. [DOI] [PubMed] [Google Scholar]
- 20.Matsuda, K., T. Hirokazu, T. Asahara, Y. Kado, and K. Nomoto. 2007. Sensitive quantitative detection of commensal bacteria by rRNA-targeted reverse transcription-PCR. Appl. Environ. Microbiol. 7332-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 977539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller, C., and A. J. Macpherson. 2006. Layers of mutualism with commensal bacteria protect us from intestinal inflammation. Gut 55276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Hara, A. M., and F. Shanahan. 2006. The gut flora as a forgotten organ. EMBO Rep. 7688-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Que, J. U., and D. J. Hentges. 1985. Effect of streptomycin administration on colonization resistance in mice. Infect. Immun. 48169-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rakoff-Nahoum, S., J. Paglino, F. Eslami-Varzaneh, S. Edberg, and R. Medzhitov. 2004. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118229-241. [DOI] [PubMed] [Google Scholar]
- 26.Rescigno, M., M. Urbano, B. Valzasina, M. Fancolini, G. Rotta, R. Bonasio, F. Granucci, J. P. Kraehenbuhl, and P. Ricciardi-Castagnoli. 2001. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2361-367. [DOI] [PubMed] [Google Scholar]
- 27.Rinttila, T., A. Kassinen, E. Malinen, L. Krogius, and A. Palva. 2004. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 971166-1177. [DOI] [PubMed] [Google Scholar]
- 28.Salzman, N. H., M. M. Chou, H. de Jong, L. Liu, E. M. Porter, and Y. Paterson. 2003. Enteric Salmonella infection inhibits Paneth cell antimicrobial peptide expression. Infect. Immun. 711109-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salzman, N. H., H. de Jong, Y. Paterson, H. J. M. Harmsen, G. W. Welling, and N. A. Bos. 2002. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology 1483651-3660. [DOI] [PubMed] [Google Scholar]
- 30.Snel, J., P. P. Heinen, H. J. Blok, R. J. Carman, A. J. Duncan, P. C. Allen, and M. D. Collins. 1995. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus Arthromitus.” Int. J. Syst. Bacteriol. 45780-782. [DOI] [PubMed] [Google Scholar]
- 31.Stecher, B., R. Robbiani, A. W. Walker, A. M. Westendorf, M. Barthei, M. Kremer, S. Chaffron, A. J. Macpherson, J. Buer, J. Parkhill, G. Dougan, C. von Mering, and W. D. Hardt. 2007. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5e244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan, M. P., M. Kaparakis, M. Galic, J. Pedersen, M. Pearse, O. L. Wijburg, P. H. Janssen, and R. A. Strugnell. 2007. Chronic Helicobacter pylori infection does not significantly alter the microbiota of the murine stomach. Appl. Environ. Microbiol. 731010-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wells, C. L., M. A. Maddaus, C. M. Reynolds, R. P. Jechorek, and R. L. Simmons. 1987. Role of anaerobic flora in the translocation of aerobic and facultatively anaerobic intestinal bacteria. Infect. Immun. 552689-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wise, M. G., and G. R. Siragusa. 2005. Quantitative detection of Clostridium perfringens in the broiler fowl gastrointestinal tract by real-time PCR. Appl. Environ. Microbiol. 713911-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wostmann, B. S. 1981. The germfree animal in nutritional studies. Annu. Rev. Nutr. 1257-279. [DOI] [PubMed] [Google Scholar]