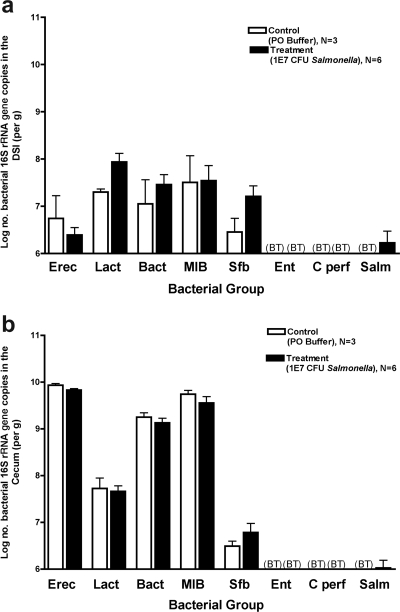
FIG. 8.
Quantitative analysis of the intestinal microbiota after intestinal clearance of S. enterica serovar Typhimurium. Mice were inoculated with 107 CFU S. enterica serovar Typhimurium and sacrificed after 30 days. The intestinal tract was removed and divided into the DSI, cecum, and LI. Bacterial genomic DNA was isolated from each segment, and qPCR analysis was performed to determine the abundance of specific commensal bacterial groups in the DSI (a) and cecum (b). White bars represent uninfected controls. Black bars represent Salmonella-infected mice. Salmonella infection did not appear to affect the bacterial numbers in any segment of the gut (P > 0.05). BT, below the detection threshold of qPCR. Erec, Eubacterium rectale/Clostridium coccoides; Lact, Lactobacillus sp.; Bact, Bacteroides sp.; MIB, mouse intestinal Bacteroides; Sfb, segmented filamentous bacteria; Ent, Enterobacteriaceae; C. perf, Clostridium perfringens; Salm, S. enterica serovar Typhimurium.