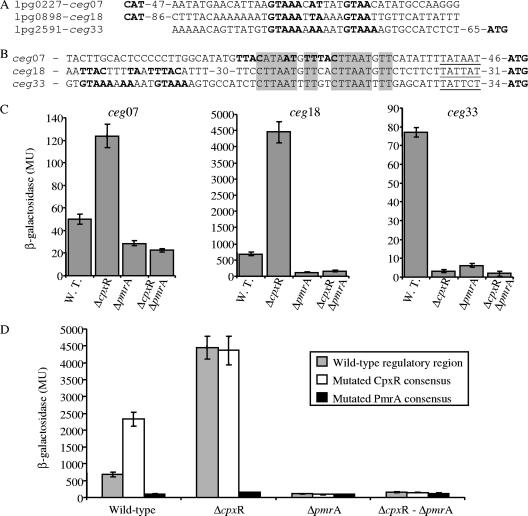
FIG. 7.
ceg genes are regulated by the CpxR and PmrA regulators. (A and B) Sequence alignment of the regulatory regions of the three ceg genes according to the CpxR consensus sequence (A) or the PmrA consensus sequence (B). The putative PmrA consensus sequence is indicated by shading, the putative CpxR consensus sequence is indicated by boldface type, the putative −10 promoter elements are underlined, and the distances to the first ATG codon are also indicated. (C) Expression of the three lacZ fusions in wild-type L. pneumophila (W.T.), cpxR deletion mutant OG2002 (ΔcpxR), pmrA deletion mutant HK-PQ1 (ΔpmrA), and cpxR pmrA double-deletion mutant EA-CRPA (ΔcpxR ΔpmrA). (D) Expression of the wild-type ceg18::lacZ fusion (grey bars), the ceg18::lacZ fusion containing a mutation in the CpxR consensus sequence (open bars), and the ceg18::lacZ fusion containing a mutation in the PmrA consensus sequence (black bars) in wild-type L. pneumophila (Wild-type), cpxR deletion mutant OG2002 (ΔcpxR), pmrA deletion mutant HK-PQ1 (ΔpmrA), and cpxR pmrA double-deletion mutant EA-CRPA (ΔcpxR-ΔpmrA). β-Galactosidase activity was determined as described in the Materials and Methods. The data are the averages ± standard deviations (error bars) of at least three different experiments. MU, Miller units.