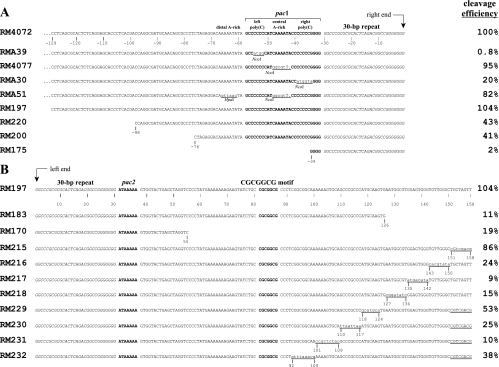
FIG. 2.
Sequences of ectopic cleavage sites. Sequences close to the point of cleavage are shown to illustrate relevant nucleotide differences between the ectopic cleavage sites of different viruses. Corresponding cleavage site efficiencies are given to the right of each sequence. (A) Substitutions and nested deletions on the pac1 side of the cleavage site with predicted right ends aligned to the right. (B) Nested deletions and scanning substitutions on the pac2 side of the cleavage site with predicted left ends aligned to the left. Known functional cis elements (pac1, pac2, and the CGCGGCG motif) are in bold and set off by spaces; 30-bp terminal repeats are in italics. Nucleotide positions are numbered negatively to the left of right-end termini and positively to the right of left-end termini. Substitution mutations are underlined, nucleotide changes are in lower case, and a dot indicates a missing base. Restriction sites created by mutations are indicated. Note that although a single 30-bp terminal repeat is shown at each terminus, pac2-containing termini sometimes lack a 30-bp repeat (16); the disposition of 30-bp repeats at ectopic termini described here has not been determined.