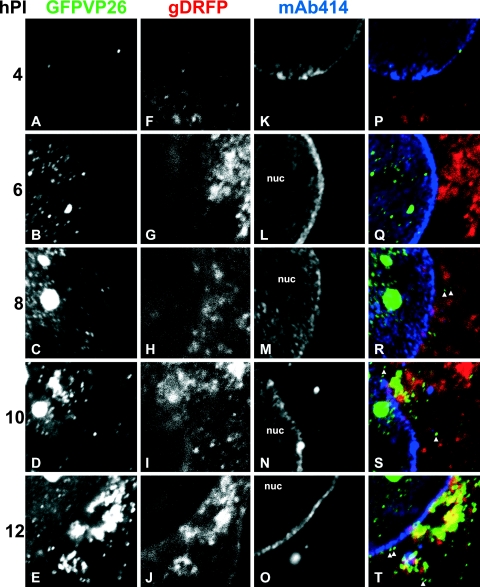
FIG. 7.
Time course of HSV1 nuclear egress and assembly. Vero cells were inoculated with 10 PFU/cell of HSV1(17+)blueLox-GFPVP26-gDRFP for 2 h at 4°C to allow efficient binding and synchronous infection. Unbound virus was removed by extensive washing, the cells were transferred to 37°C, and fixed at 4 h p.i (A, F, K, and P), 6 h p.i (B, G, L, and Q), 8 h p.i (C, H, M, and R), 10 h p.i (D, I, N, and S), or 12 h p.i (E, J, O, and T). GFPVP26 (A to E; green areas in panels P to T) and gDRFP (F to J; red areas in panels P to T) were detected by their intrinsic fluorescence. After permeabilization, the cells were labeled with MAb 414 against nucleoporins (K to O; blue areas in panels P to T) and analyzed by confocal laser scanning microscopy. To evaluate the relative subcellular localization of capsids (GFPVP26), viral envelope proteins (gDRFP), and nuclear pores (MAb 414), the signals were overlaid and displayed in color (P, Q, R, S, and T). The nuclei (nuc) and some cytoplasmic capsids, which do not colocalize with gD and thus appear green in the overlays (arrowheads in panels Q and R), are indicated. The specimens were analyzed by confocal laser scanning microscopy.