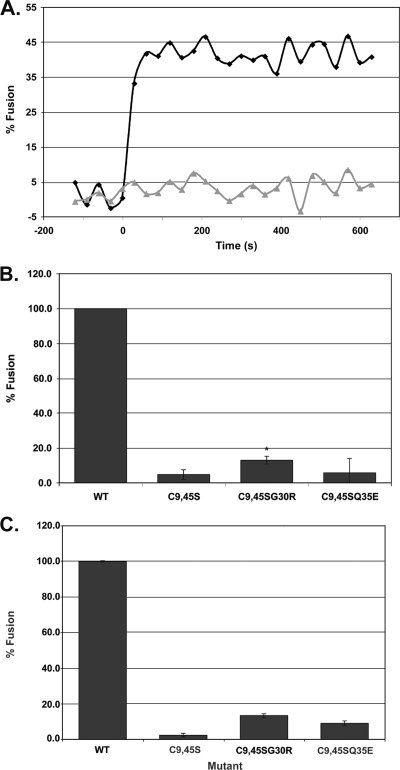
FIG. 3.
Characterization of lipid mixing induced by VSV-EnvAC9,45S and two second-site revertants. (A) Lipid mixing induced by VSV-EnvA (diamonds) and VSV-EnvAC9,45S (triangles). (B and C) Extent of lipid mixing for VSV-EnvA (wild type [WT]), VSV-EnvAC9,45S, VSV-EnvAC9,45SG30R, and VSV-EnvAC9,45SQ35E. Virus samples were prepared and lipid mixing assessed as described for Fig. 2, except that the relative amount of each virus used per assay was normalized for the relative incorporation of the various EnvA's (ranges, 0.63 to 1.42 for EnvAC9,45S, 0.63 to 0.86 for EnvAC9,45SG30R, and 0.71 to 1.02 for EnvAC9,45SQ35E, relative to wild-type EnvA) and the fusion observed for wild-type EnvA in the presence of R99 was used as pH0. Data in panel B are averages from four experiments, each carried out in triplicate. t tests were performed using the “two sample assuming unequal variances” function in Microsoft Excel [*, P(T ≤ t) = 0.0037]. Data in panel C are averages from duplicate samples from a single experiment. In panels B and C, FpH was calculated as for Fig. 2C.