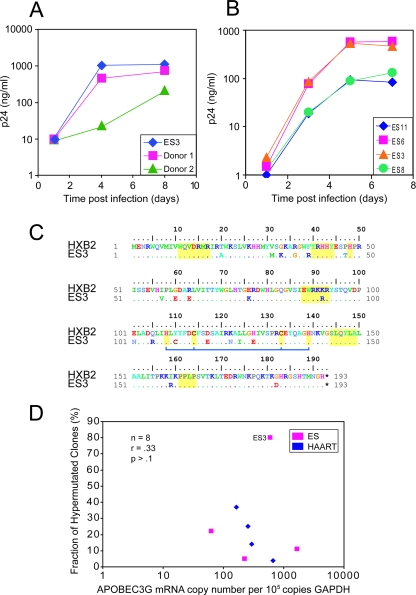
FIG. 3.
Characterization of ES3. Susceptibility of CD4+ T cells from ES3 to spreading HIV-1 infection. CD4+ T lymphoblasts from ES3 and from two normal controls (donors 1 and 2) (A) and three ESs (ES5, ES8, and ES11) (B) were isolated and infected with Ba-L. Virus growth was measured by quantitating HIV-1 p24 antigen in the culture supernatant. (C) Sequence of the vif gene from ES3. No novel substitutions, insertions, or deletions were found. There were no significant changes at sites that are known to be important in Vif functionality (regions indicated in yellow). The blue bar denotes amino acids that constitute a zinc binding motif important for APOBEC3G degradation (28, 33, 39). (D) Expression of APOBEC3G mRNA was measured using a real-time PCR assay in four ESs (ES3, ES5, ES8, and ES11) and four HAART-treated patients (patients 99, 136, 147, and 148). mRNA levels were calculated as copies of APOBEC3G per 100,000 copies of GAPDH. No correlation between the fraction of hypermutated clones and expression of APOBEC3G was seen.