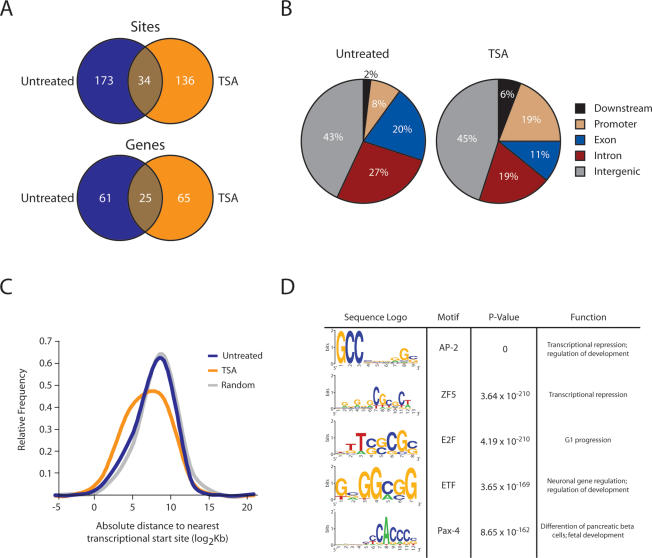
Figure 3.
Nup93 association with the genome is significantly altered upon loss of histone deacetylation. (A) Venn diagrams showing the overlap between untreated and TSA-treated Nup93-binding sites and nearest genes. (B) Comparison of untreated and TSA-treated Nup93-binding sites with chip5 distributions reveals significant changes in association across five genomic regions: exons, introns, promoters, downstream elements, and intergenic regions (P = 0.001; χ2). Untreated sites are enriched for introns when compared with chip5 (P = 0.002). TSA-binding sites are significantly enriched for introns and promoters (P = 0.005 and P = 1.2 × 10−14, respectively) and depleted for exons and intergenic regions (P = 5.7 × 10−4 and P = 5.4 × 10−5, respectively) when compared with chip5 distributions. (C) Distribution of distances from the center of each untreated Nup93-binding site, TSA-treated Nup93-binding site, and random site to the nearest transcription start site. P-values for C were calculated by comparing untreated and TSA-treated Nup93-binding sites to randomly distributed sites using the two-tailed Mann-Whitney U-test. (D) Sequence logos for the five most significantly enriched transcription factor-binding motifs identified in TSA-treated Nup93-binding sites. One-hundred-six motifs were identified in TSA-treated binding sites by CEAS (Supplemental Table 3).