Abstract
Understanding the role of transcription factors (TFs) is essential in reconstructing developmental regulatory networks. The plant-specific GeBP TF family of Arabidopsis thaliana (Arabidopsis) comprises 21 members, all of unknown function. A subset of four members, the founding member GeBP and GeBP-like proteins (GPL) 1, 2, and 3, shares a conserved C-terminal domain. Here we report that GeBP/GPL genes represent a newly defined class of leucine-zipper (Leu-zipper) TFs and that they play a redundant role in cytokinin hormone pathway regulation. Specifically, we demonstrate using yeast, in vitro, and split-yellow fluorescent protein in planta assays that GeBP/GPL proteins form homo- and heterodimers through a noncanonical Leu-zipper motif located in the C-terminal domain. A triple loss-of-function mutant of the three most closely related genes gebp gpl1 gpl2 shows a reduced sensitivity to exogenous cytokinins in a subset of cytokinin responses such as senescence and growth, whereas root inhibition is not affected. We find that transcript levels of type-A cytokinin response genes, which are involved in the negative feedback regulation of cytokinin signaling, are higher in the triple mutant. Using a GPL version that acts as a constitutive transcriptional activator, we show that the regulation of Arabidopsis response regulators (ARRs) is mediated by at least one additional, as yet unknown, repressor acting genetically downstream in the GeBP/GPL pathway. Our results indicate that GeBP/GPL genes encode a new class of unconventional Leu-zipper TF proteins and suggest that their role in the cytokinin pathway is to antagonize the negative feedback regulation on ARR genes to trigger the cytokinin response.
Transcription factors (TFs) are key regulators of developmental processes and the complexity of living organisms necessitates a large number of TFs. In plants, TFs are often involved in the control of hormone pathways and several recent studies in Arabidopsis (Arabidopsis thaliana) provide new insight into how TFs and phytohormones interact to control plant development (Long and Benfey, 2006; Shani et al., 2006). However, only a small portion of plant TFs have been functionally characterized by mutation analysis (Riechmann et al., 2000). We previously described the GeBP (GLABROUS1 enhancer-binding protein) gene, which is the founding member of a new plant-specific Arabidopsis TF family (http://datf.cbi.pku.edu.cn/browsefamily.php?familyname=GeBP) whose members share a central DNA-binding domain. None of the 21 members of the GeBP family has been assigned to a biological function.
Among these TFs, GeBP and the three GeBP-like (GPL) 1, 2, and 3 proteins form a distinct clad and share an additional C-terminal conserved region of unknown function (Curaba et al., 2003). GeBP is predicted to play a role in hormonal pathways on the basis of the following observations: (1) the GeBP protein binds the cis-regulatory element of the GLABROUS1 gene, a myb-gene regulated by GA and cytokinin hormones (Perazza et al., 1998; Gan et al., 2007) involved in epidermis cell determination (Oppenheimer et al., 1991); and (2) transcript levels of GeBP are positively regulated by BREVIPEDICELLUS (BP; Curaba et al., 2004), a gene of the KNOTTED1 homeodomain (KNOX) family that positively regulates the cytokinin pathway in the shoot apical meristem (SAM; Jasinski et al., 2005; Yanai et al., 2005). Hormones such as cytokinins, GAs, and auxin are involved in the establishment of the balance between the production of organs from the flanks of the SAM and indeterminate growth at its center. Cytokinins, which positively regulate cell division (Riou-Khamlichi et al., 1999; Howell et al., 2003; Ferreira and Kieber, 2005), are required for meristem function and maintenance (Giulini et al., 2004; Leibfried et al., 2005; Kurakawa et al., 2007). GAs and auxin act antagonistically to cytokinins with GAs promoting cell differentiation (Chien and Sussex, 1996; Ogas et al., 1997; Perazza et al., 1998; Hay et al., 2002) and auxin promoting organ initiation (Reinhardt et al., 2000). Cytokinin signaling in plants is similar to the bacterial two-component phosphorelay system composed of a His kinase sensor and a response regulator. The cytokinin receptors that have a receiver domain fused to a His kinase domain are predicted to signal through His phosphotransfer proteins to ultimately alter the phosphorylation of response regulators (Hutchison and Kieber, 2002). The Arabidopsis response regulators (ARRs) are classified as type A or type B based on their sequence similarities. The rate of transcription of most type-A ARRs, but not of type-B ARRs, is rapidly and specifically induced in response to exogenous cytokinin (D'Agostino et al., 2000) and is dependent at least in part on type-B ARRs (Hwang and Sheen, 2001; Sakai et al., 2001). Importantly, in contrast with type-B ARRs, type-A ARRs are involved in negative feedback regulation of cytokinin signaling (Kiba et al., 2003; To et al., 2004; Hutchison et al., 2006).
We show here that the four GeBP/GPL genes encode a newly defined class of unconventional Leu-zipper proteins and are involved in cytokinin response regulation. This regulation is shown by the finding that a triple loss-of-function mutant is less sensitive to exogenous cytokinin and that transcript levels of type-A ARR cytokinin response genes are increased, likely resulting in an increased negative feedback regulation and ultimately cytokinin insensitivity. Conversely, overexpression of a GPL protein with a constitutive transcriptional-activation activity causes a decrease in type-A ARR transcript levels together with an increased sensitivity to cytokinins, indicating that the GeBP/GPL-dependent regulation of ARRs involves additional unknown repressing TFs acting downstream of GeBP/GPLs. Taken together, these results suggest that the role of GeBP/GPL genes in cytokinin signaling is to antagonize the negative feedback regulation by repressing type-A ARRs through the action of one or several repressors that remain to be identified.
RESULTS
GeBP Family Members Form Homo- and Heterodimers
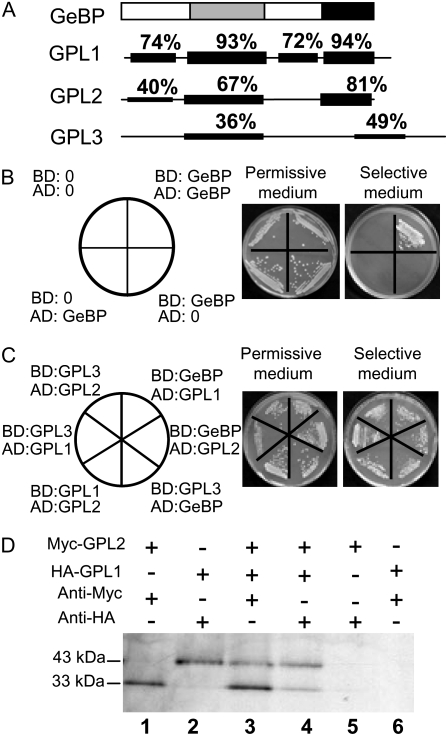
Previous work on GeBP led to the definition of a new GeBP gene family in Arabidopsis with 21 members, all of unknown function (Curaba et al., 2003). A subset of four members, namely GeBP and GPL proteins 1, 2, and 3, share two plant-specific conserved regions (Fig. 1A): a central domain homologous to the DNA-binding domain of the STORE-KEEPER (STK) TF from potato (Solanum tuberosum; Zourelidou et al., 2002) and a C-terminal region of unknown function (Curaba et al., 2003). Both domains were shown to be necessary for trans-activation of reporter genes in yeast one-hybrid experiments (Curaba et al., 2003). This suggested that GeBP could bind DNA through its central domain while the C-terminal region could stabilize this activity possibly by forming homodimers. To test this dimerization hypothesis, GeBP was fused to the activation domain (AD) or binding domain (BD) of the Gal4 TF and the fusion constructs were cotransformed into yeast. As shown in Figure 1B, cotransformants were able to grow on selective medium, indicating that the full-length GeBP forms homodimers in yeast. Similarly, GPL1, GPL2, and GPL3 were fused to the AD and the BD and were also shown to homodimerize in two-hybrid experiments (data not shown). The formation of heterodimers was then tested by cotransforming yeast with all possible pairs of GeBP family members. The six combinations all enabled yeast to grow on selective medium (Fig. 1C), indicating that all heterodimers can form in yeast even between GeBP and GPL3, the two most divergent proteins. This property of GeBP family members to form heterodimers was further tested by in vitro immunoprecipitation. 35S-labeled GPL1 and GPL2 tagged with HA and Myc epitopes, respectively, were synthesized in vitro using rabbit reticulocyte lysate. Incubation with either anti-HA or anti-Myc antibodies led to the specific coimmunoprecipitation of both proteins (Fig. 1D). These experiments indicate that all combinations of homo- and heterodimers can be formed between the four GeBP/GPL proteins.
Figure 1.
Dimerization of GeBP and GPL proteins. A, Schematic representation of the GeBP protein and its similarities with GPL proteins. Gray and black areas represent the conserved DNA-binding and C-terminal domains, respectively. B, Homodimerization assay of GeBP in yeast. C, Heterodimerization assay of GeBP and GPL proteins in yeast. D, Coimmunoprecipitation assay of GPL1 and GPL2 proteins in vitro. Myc-tagged GPL2 and HA-tagged GPL1 were translated separately in vitro in the presence of [35S]Met and immunoprecipitated independently with the corresponding anti-tag antibodies. Translation mixes were combined in a 1:1 ratio (lane 3 and lane 4). Immunoprecipitation with the anti-myc antibody (lane 1, lane 3, and lane 6) or with the anti-HA antibody (lane 2, lane 4, and lane 5).
The C-Terminal Region of GeBP and GPL Proteins Harbors a Functional Noncanonical Leu-Zipper Motif
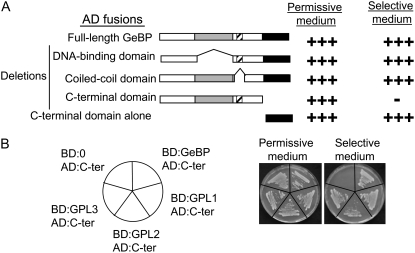
A series of deletions were made in the AD:GeBP fusion protein to determine which region of the protein was involved in dimer formation. These deletions covered three regions: the DNA-binding domain, a predicted coiled-coil region (often involved in protein-protein interaction), and the C-terminal domain (Fig. 2A). As expected, the deletion of the DNA-binding domain did not prevent yeast growth. Similarly, deletion of the coiled coil did not prevent yeast growth, indicating that this region is not involved in dimer formation. On the contrary, deletion of the conserved C-terminal region completely abolished yeast growth, indicating that this region is necessary for dimer formation. Conversely, the C-terminal region alone trans-activated yeast reporter genes, showing that this region is sufficient for GeBP dimerization (Fig. 2A). Cotransformation of just the GeBP C-terminal region with full-length GPL1, 2, or 3 also led to yeast growth (Fig. 2B). Therefore the C-terminal region of the GeBP protein is responsible for homo- and heterodimer formation.
Figure 2.
Mapping of the GeBP dimerization domain. A, Internal deletions within the wild-type GeBP protein. Deletions were made in the AD:GeBP fusions and cotransformed with the wild-type BD:GeBP fusion. Gray, hatched, and black areas represent the DNA-binding domain, putative coiled-coil region, and C-terminal domain, respectively. B, Assay of the interaction between the GeBP C-terminal domain and GPL proteins in yeast.
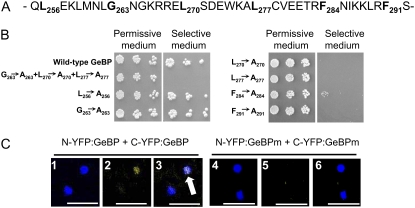
A putative partial Leu-zipper motif in the C-terminal domain of GeBP/GPL proteins was described previously (Curaba et al., 2003). Conventional Leu-zipper motifs have four to seven Leu residues, each separated by six amino acids (Bornberg-Bauer et al., 1998). The putative GeBP motif (Fig. 3A) consists of six residues (Leu-256, Gly-263, Leu-270, Leu-277, Phe-284, and Phe-291), three of which are non-Leu residues. We tested whether this putative partial Leu-zipper motif was responsible for GeBP dimerization by mutating specific amino acids. First, we simultaneously mutated the three central amino acids Gly-263, Leu-270, and Leu-277 into Ala residues. This mutated version of GeBP fused to the AD was cotransformed in yeast with the wild-type GeBP fused to the BD. As shown on Figure 3B, with this combination yeast growth was completely suppressed, suggesting that the wild-type motif was functional. We next tested the functionality of all six residues (Leu-256, Gly-263, Leu-270, Leu-277, Phe-284, and Phe-291) by mutating them individually to Ala residues. When each of the first two residues, Leu-256 and Gly-263, was mutated individually, yeast growth was not affected, indicating that these residues do not play a role in dimer formation. On the contrary, when each of the four remaining residues, Leu-270, Leu-277, Phe-284, and Phe-291, was mutated individually, yeast growth was abolished (Fig. 3B). These results indicate that only the last four residues of the putative Leu-zipper motif are essential for dimer formation. We concluded that GeBP can form dimers through the following noncanonical Leu-zipper motif: Leu-270(X)6Leu-277(X)6Phe-284(X)6Phe-291.
Figure 3.
Functionality of the noncanonical Leu-zipper motif. A, Primary sequence of the putative Leu-zipper motif of GeBP. Putative residues of the Leu-zipper motif are shown in bold. B, Mutagenesis of the putative Leu-zipper motif. Drop (5 μL) test of serial 5-fold dilutions (starting at A600 = 0.2) of a permissive medium liquid culture on permissive and selective plates. Residues of the putative Leu-zipper motif that were changed to Ala residues are indicated. C, BiFC with wild-type and mutated full-length GeBP in plant cells. Tobacco leaves were coinfiltrated with N-YFP:GeBP and C-YFP:GeBP (1–3) or N-YFP:GeBPm and C-YFP:GeBPm (4–6). The mutated version has the three mutations (Gly-263, Leu-270, and Leu-277) described in Figure 3B. 1 and 4, DAPI signal showing nuclei; 2 and 5, YFP channel; 3 and 6, merging of DAPI and YFP signals where white spots (white arrow) indicate colocalization of DAPI and YFP signals. Scale bars, 50 μm.
To confirm that these interactions occur in plant cells, we tested dimer formation using the bimolecular fluorescence complementation (BiFC) technique, which allows protein-protein interactions to be visualized in situ (Walter et al., 2004). Using Agrobacterium tumefaciens transformation, tobacco (Nicotiana benthamiana) leaf cells were cotransformed with two constructs: one encoding a fusion between the N-terminal half of yellow fluorescent protein (YFP) and GeBP, and the other encoding a fusion between the C-terminal half of YFP and GeBP. Nuclei of leaf cells were stained with 4′,6′-diamidino-2-phenylindole hydrochloride (DAPI; Fig. 3C). The GeBP protein is known to be localized in the nucleus (Curaba et al., 2003). YFP fluorescence was detected in epidermal cells and colocalized with the DAPI staining (Fig. 3C). As a control, the GeBP mutant version with three residues mutated in the Leu-zipper was fused to the two halves of YFP and the two constructs were cotransformed in plant cells as reported above. No YFP signal was visible in DAPI-stained nuclei (Fig. 3C). GeBP and GPL2 were also tested and heterodimerization was observed (data not shown). These data indicate that GeBP/GPLs can form dimers in vivo and demonstrate that these proteins are a new unusual class of Leu-zipper TFs.
GeBP/GPL Genes Encode Nuclear Proteins and Display Overlapping Expression Patterns
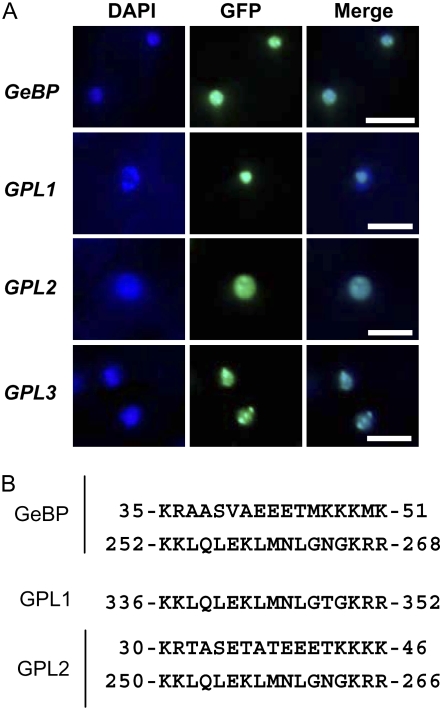
To determine the intracellular localization of the GeBP/GPL proteins, the four corresponding coding sequences were cloned downstream of and in frame with the GFP reporter gene under the control of the constitutive 35S promoter. In transgenic lines of Arabidopsis, the four GFP fusion proteins were localized in nuclei (Fig. 4A). This is consistent with the presence of at least one nuclear localization signal (NLS) in GeBP, GPL1, and GPL2 (Fig. 4B). The GPL3 protein was also localized in nuclei despite the lack of an obvious NLS in its primary sequence.
Figure 4.
Intracellular localization and motifs of GeBP and GPL proteins. A, Subcellular localization of GeBP/GPL proteins. Stable Arabidopsis transgenic lines were transformed with a binary vector containing GFP:GeBP/GPL fusion constructs under the control of the 35S promoter. Epidermal cells were stained with DAPI to visualize nuclei and observed under epifluorescence microscopy using a DAPI filter or a GFP filter. DAPI and GFP images were merged to show the colocalization of both signals. Scale bars, 20 μm. B, Predicted NLSs in GeBP, GPL1, and GPL2 proteins. GPL3 protein has no obvious NLS.
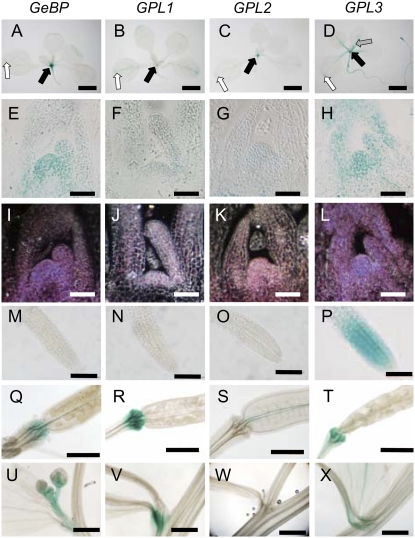
Spatial and temporal expression of the GeBP and GPL genes was further examined by generating lines carrying Promoter:GUS (P:GUS) fusions for each gene. During vegetative development, the PGeBP:GUS, PGPL1:GUS, PGPL2:GUS, and PGPL3:GUS reporter lines showed largely overlapping expression patterns with the main expression being in the SAM and young leaf primordia (Fig. 5, A–L). The strongest staining was observed in GeBP lines and the weakest in the GPL1 lines, these two genes being the two most similar homologs. The vascular tissues of cotyledons and leaves and hydathodes also stained weakly in the GeBP, GPL1, and GPL2 lines (Fig. 5, A–D). No GUS staining was observed in roots of the GeBP, GPL1, and GPL2 reporter lines (Fig. 5, M–O). In contrast, the GPL3 reporter lines showed a strong staining of primary and secondary roots (Fig. 5P) as well as a marked staining of vascular tissues of rosette leaves (Fig. 5D). During reproductive development, the four GeBP/GPL reporter lines still had overlapping expression patterns with GUS staining in the distal part of pedicels that form a vascular bulge at the base of flowers and siliques (Fig. 5, Q, R, and T), except GPL2 lines where the staining was localized in the septum of siliques (Fig. 5S). The paraclades were also frequently stained in all lines analyzed (Fig. 5, U, V, and X), except GPL2 lines for which no staining was observed (Fig. 5W). Overall, we conclude that GeBP/GPL genes have largely overlapping expression patterns and are mainly expressed in the SAM, young leaf primordia, and vascular tissues.
Figure 5.
Expression analysis of GeBP/GLP promoters. A to D, Staining of 15-d-old rosettes. Black arrows, SAM staining; white arrows, hydathode staining; gray arrow, vasculature staining. Scale bars, 2 mm. E to H, Cross sections of rosette-stage SAMs. Image contrast was increased using GIMP software (F). Scale bars, 50 μm. I to L, Dark-field illumination of cross sections shown to visualize weak GUS staining. Scale bars, 50 μm. M to P, Staining of a primary root meristems. Scale bar, 100 μm. GeBP, GPL1, and GPL2 reporter lines show no staining in primary roots (M to O) in contrast to GPL3 lines (P). Q to T, Staining of silique pedicels. Scale bars, 500 μm. U to X, Staining of paraclades. GPL2 expression is limited to the septum. Scale bars, 1 mm.
The Triple Mutant gebp-1 gpl1-1 gpl2-2 Shows a Reduced Sensitivity to Exogenous Cytokinins
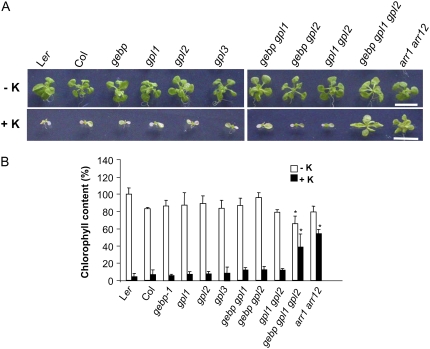
The overlapping expression patterns of the GeBP/GPL genes and their ability to interact in all combinations at the protein level are suggestive of functional redundancy. To study this redundancy, single mutants corresponding to the three most closely related genes, GeBP, GPL1, and GPL2, were isolated (Supplemental Fig. S1) and crossed to construct all the corresponding double mutants and the triple mutant. Although the double mutants were indistinguishable from wild types, the triple mutant gebp-1 gpl1-1 gpl2-1 was slightly paler than the wild types when grown on Murashige and Skoog (MS) medium (see below). As the founding member GeBP acts downstream of KNOXI genes (Curaba et al., 2003) whose main role is to control hormonal pathways, especially GA and cytokinin pathways (Sakamoto et al., 2001; Chen et al., 2004; Jasinski et al., 2005; Yanai et al., 2005), the triple mutant was grown in the presence of different hormones, including GAs, cytokinins, abscisic acid, auxins, the ethylene precursor 1-aminocyclopropane-1-carboxylic acid, jasmonic acid, and brassinosteroids. When grown on MS medium supplemented with various concentrations of each of these hormones, the triple mutant was indistinguishable from the wild types (data not shown) except with cytokinin hormones (Fig. 6A). In the presence of cytokinins such as kinetin, wild-type plants germinated normally but soon stopped growing and failed to develop shoots, as has been described previously (Higuchi et al., 2004; To et al., 2004; Hutchison et al., 2006; Fig. 6A). Similarly to wild types, single and double gebp/gpl mutants were also severely affected by cytokinins although cotyledons stayed green for longer and plants sometimes initiated true leaf primordia before growth ceased. In contrast, the development of the triple mutant did not arrest in the presence of exogenous cytokinins and the shoot developed visible leaves that remained green (Fig. 6A). The type-B ARR gene double mutant, arr1-3 arr12-1, which is impaired in cytokinin signaling (Mason et al., 2005), was included and was markedly less sensitive to cytokinins than wild type in our assay, as expected (Fig. 6A). In cytokinin root inhibition assays and lateral root formation assays, single, double, and triple mutants were indistinguishable from wild types (data not shown), consistent with the absence of expression in roots of the GeBP, GPL1, and GPL2 reporter lines. This suggests that GeBP/GPL genes play a role in cytokinin responses in a subset of organs or tissues rather than in the whole plant as described for cytokinin signaling (Ferreira and Kieber, 2005).
Figure 6.
Sensitivity of gebp, gpl1, and gpl2 mutants to cytokinin. A, In vitro growth of wild types and single, double, and triple mutants on MS in the absence (−K) or presence (+K) of kinetin (10 μg mL−1). The triple mutant was distinguishable from wild type on lower concentrations (2–5 μg mL−1) of kinetin and was observed on trans-zeatin (1 μg mL−1) as previously described (Higuchi et al., 2004; To et al., 2004; Hutchison et al., 2006). Scale bars, 1 cm. B, Chlorophyll content of wild types and single, double, and triple mutants grown in the absence (white bars) or presence (black bars) of kinetin. Asterisks represent significant changes to corresponding wild-type controls in the presence or absence of kinetin using the ANOVA test. Plants were grown for 20 d. The value for wild-type Ler was set at 100%. The experiment was done at least three times with consistent results. [See online article for color version of this figure.]
Because cytokinins play a role in leaf chlorophyll content (Richmond and Lang, 1957; Gan and Amasino, 1995), the chlorophyll content of wild types and mutants grown in the absence or presence of cytokinins was quantified (Fig. 6B). In the absence of cytokinins, the triple gebp/gpl mutant showed the lowest chlorophyll content relative to single and double mutants and wild types. In the presence of cytokinins, chlorophyll contents were severely reduced in wild types and single mutants and to a lesser extend double mutants. In contrast, chlorophyll contents in both the double arr mutant and the triple gebp/gpl mutant showed only a mild reduction. Altogether, these data indicate that the triple mutant had a reduced sensitivity to cytokinins in aerial organs and showed a phenotype similar to the arr1 arr12 double mutant, suggesting a decreased cytokinin pathway in the triple mutant.
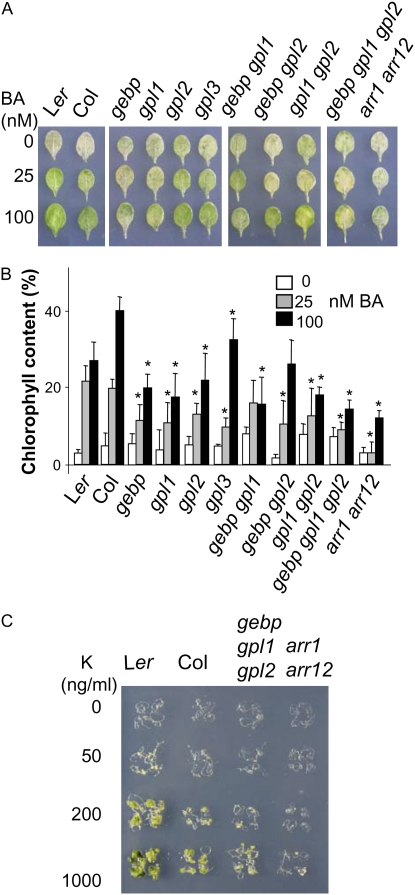
To further investigate the effect of GeBP/GPL mutations on the response of aerial parts of the plant to cytokinins, we measured the effect of cytokinins on detached leaves during the process of dark-induced senescence, which partially mimics senescence processes, including chlorophyll degradation (Ueguchi et al., 2001; Buchanan-Wollaston et al., 2005). After 10 d of dark-induced senescence, wild-type leaf chlorophyll levels fell substantially (Fig. 7, A and B). This decrease in chlorophyll levels was inhibited by cytokinins such as 6-benzyl-adenine (BA). Chlorophyll levels also fell in the double mutant arr1 arr12 in the absence of cytokinin, and as expected this decrease was not suppressed in the presence of cytokinin, contrasting with the wild-type response. In single, double, and triple gebp/gpl mutants in the absence of cytokinin, the chlorophyll content decreased to varying extents. With cytokinin, the decrease in chlorophyll content of most mutants was greater relative to the wild-type response, indicating that gebp/gpl mutants had largely lost their ability to retain chlorophyll in response to cytokinin treatment. Single and double mutants were more distinguishable from wild-type controls in the dark-induced senescence assay than in the growth assay, suggesting that the dark-induced senescence assay was a more sensitive way of measuring the cytokinin response. Furthermore, we cannot exclude that the two different genetic backgrounds may also contribute to the observed phenotype in these assays.
Figure 7.
Dark-induced senescence in a detached leaf assay and its inhibition by cytokinin. A, Detached leaves of wild types and mutants incubated for 10 d in the dark in the presence of the indicated BA concentrations. B, Quantification of chlorophyll content in detached leaves. The leaf chlorophyll content before the start of dark incubation was set at 100% for each genotype tested. Asterisks represent significant changes to wild-type controls at respective hormone concentrations using the ANOVA test. Results shown are pooled from three independent experiments. C, Callus induction assays. Root segments were incubated in the absence or presence of kinetin (K) at the indicated concentrations. Four representative root segments are shown for each genotype at each hormone concentration.
Cytokinins normally stimulate cell division and greening of calli. According to microarray databases, at least GeBP and GPL1 are strongly expressed in calli. Therefore, responses of tissue-cultured explants were examined with varied cytokinin concentrations in the presence of the auxin 2,4-dichlorophenoxyacetic acid (Fig. 7C). After 3 weeks, segments of wild-type roots responded by forming green calli at the two highest cytokinin concentrations tested, whereas the arr1 arr12 double mutant did not, as has been described previously (Mason et al., 2005). The triple mutant formed less calli that remained yellow, indicating that it was less responsive to exogenous cytokinins.
Taken together, these data show that the GeBP, GPL1, and GPL2 genes play redundant roles in cytokinin responses.
Transcript Levels of Type-A ARR Genes Are Up-Regulated in gebp/gpl Mutants
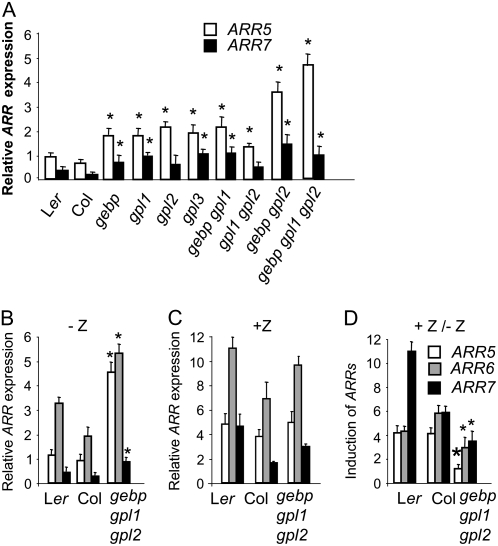
Cytokinin receptors are predicted to signal through His-phosphotransfer proteins to alter the phosphorylation of ARRs (Hutchison and Kieber, 2002). Type-A ARRs are considered to be primary cytokinin response genes and their transcription is rapidly elevated in response to exogenous cytokinin (D'Agostino et al., 2000; Kiba et al., 2002; Rashotte et al., 2003). Analyses of type-A overexpressing or high-order loss-of-function mutants indicate that type-A ARRs are redundant negative regulators of cytokinin signaling (Taniguchi et al., 1998; D'Agostino et al., 2000; Hwang and Sheen, 2001; To et al., 2004). Furthermore, type-A genes such as ARR5 and ARR6 are mostly expressed in the SAM of rosettes or pedicel bulges in a pattern that resembles GeBP/GPL expression patterns (D'Agostino et al., 2000; To et al., 2004). The reduced sensitivity of the gebp/gpl mutants could be due either to diminished cytokinin biosynthesis/signal transduction or to an increased negative feedback on cytokinin signaling. We first measured transcript levels of two ARRs genes, ARR5 and ARR7, in rosettes of wild types, and single, double, and triple mutants (Fig. 8, A and B). Although the wild types had basal levels of ARR transcripts, all mutants had higher levels of both the ARR transcripts, indicating that cytokinin feedback regulation was increased when GeBP/GPL function was impaired. The largest difference in expression was measured for ARR5 whose transcript level was 5- to 6-fold higher in the triple mutant than in wild types. Among single and double mutants, the gebp gpl2 mutant had the strongest misregulation of both ARRs. These data indicated that the GeBP/GPL genes have redundant roles in the regulation of ARR transcript levels.
Figure 8.
Transcript levels of ARR cytokinin response genes in gebp/gpl mutants. RT-PCR experiments from whole rosettes were performed in exponential amplification conditions and average integrated density ratios of ARR/Actin8 signals were determined (see “Materials and Methods”). A, Relative expression levels of ARR5 and ARR7 in wild types and single, double, and triple mutants in the absence of cytokinin. B to D, Relative expression levels of ARR5, ARR6, and ARR7 in wild types and triple mutants. Plants were grown in the absence (B) or presence (1 h) of exogenous trans-zeatin (C) and the fold inductions are shown (D). Bars in B and C are like indicated in D. Results in B were confirmed using real-time RT-PCR for ARR5. Asterisks represent significant changes to wild-type controls using the ANOVA test. Results shown are pooled from three independent experiments.
We next asked whether the triple mutant was still responsive to exogenous cytokinin relative to wild types by measuring transcript levels of three ARRs in the absence and presence of the cytokinin trans-zeatin (Fig. 8, C and D). In the absence of cytokinin, all ARR transcript levels were higher in the triple mutant than in wild types. Upon exogenous cytokinin treatment, levels of the three ARR transcripts increased in the wild types within 1 h (Fig. 8, C and D) with ARR7 being induced the most. In the triple mutant, ARR transcript levels also increased but cytokinin induction was weaker than in wild type for all three ARR genes. Thus ARR induction in the triple mutant was partially insensitive to exogenous cytokinin treatment, indicating that the triple mutant has a reduced responsiveness to exogenous cytokinins.
Overall we conclude that transcript levels of type-A ARR response genes are regulated redundantly by the GeBP, GPL1, and GPL2 genes.
GeBP/GPL Genes Regulate ARR Expression through an Indirect Repressing Pathway
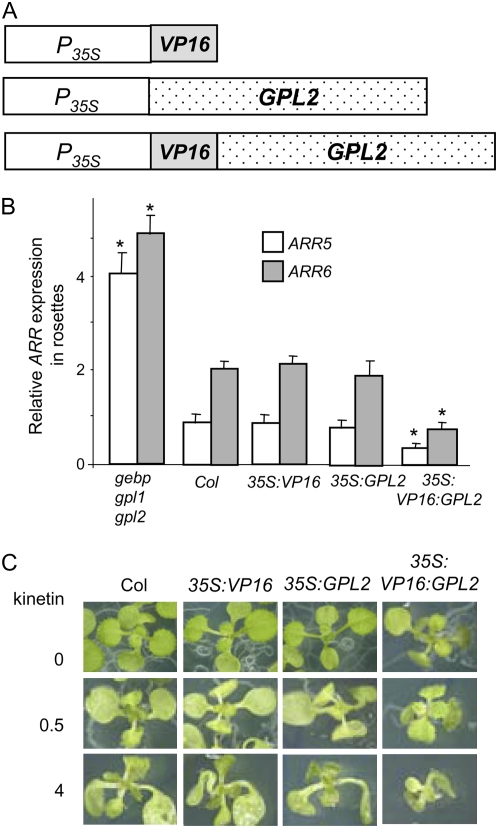
To distinguish between a direct or indirect repression of ARR gene expression by GeBP/GPLs, we generated a version of GPL2 with a constitutive transcriptional-activation activity. In this version, GPL2 is fused to the strong AD from the viral TF VP16 (Parcy et al., 1998). The rationale is that an up-regulation of ARR expression by VP16:GPL2 might indicate a direct control of ARR by GPL2, whereas a down-regulation implies an indirect control. Transgenic plants carrying a 35S:VP16:GPL2 fusion as well as 35S:VP16 or 35S:GPL2 control constructs (Fig. 9A) were obtained. In rosettes of 35S:VP16:GPL2 plants, transcript levels of ARRs were clearly lower than in rosettes of 35S:VP16, 35S:GPL2, or wild-type control plants (Fig. 9B). This indicates that the VP16:GPL2 fusion activates an unknown repressor that decreases ARR expression. Therefore the regulation of ARR is mediated by at least one unknown repressor acting genetically downstream in the GPL2 pathway. This result strongly reinforces the regulation of ARRs by GeBP/GPL genes as potential bias such as secondary mutations or ecotype backgrounds in gebp/gpl mutants are absent in 35S:VP16:GPL2 transgenic lines.
Figure 9.
Transcript levels of ARR cytokinin response genes in rosettes and cytokinin sensitivity assay in 35S:VP16:GPL2 plants. A, Schematic representation of constructs used. 35S promoter (P35S), VP16 AD (VP16), and GPL2 cDNA (GPL2) are represented. B, RT-PCR experiments were performed and quantified as in Figure 8. Asterisks represent significant changes to wild-type control using the ANOVA test. The experiment was done at least three times with consistent results and results were confirmed by real-time RT-PCR for ARR5. C, Growth inhibition by exogenous cytokinins. Wild-type and transgenic lines were grown on MS plates supplemented with the indicated concentrations of kinetin (μg mL−1) for 14 d. Several transgenic lines were tested and gave the same results. [See online article for color version of this figure.]
To determine the cytokinin sensitivity of the 35S:VP16:GPL2 transgenic lines, plants were grown in the presence of exogenous cytokinins (Fig. 9C). Rosettes of 35S:VP16:GPL2 plants exhibited a reduced growth relative to 35S:VP16, 35S∷GPL2, or wild-type rosettes and were therefore more sensitive to exogenous cytokinin (Fig. 9C). This result is consistent with an increased cytokinin signaling due to the reduction in ARR transcript levels. Similarly, in root inhibition assays, 35S:VP16:GPL2 plants exhibited shorter roots relative to 35S:VP16, 35S∷GPL2, or wild-type roots and were therefore more sensitive to exogenous cytokinin (data not shown). These results strongly support the role of GeBP/GPL genes in the regulation of cytokinin response genes.
DISCUSSION
Here we have characterized GeBP family members in Arabidopsis whose functions were unknown. A set of molecular and genetic tools were used to dissect the role of the GeBP and GPL genes in Arabidopsis development.
GeBP/GLP Proteins Are Noncanonical Leu-Zipper TFs
Proteins cannot be assigned to functional categories solely on the basis of sequence similarity to proteins or domains of known function. The GeBP/GPL proteins have not been classified as Leu-zipper TFs in databases. Leu-zipper motifs can be defined as coiled coils consisting of four to seven repeats of seven amino acids denoted a to g (Mason et al., 2006). Residues a and d consist largely of hydrophobic residues with Leus very often found in d positions (Bornberg-Bauer et al., 1998), whereas residues at e and g positions are charged. The GeBP motif identified in our work matches this definition at all d and g positions but differs mainly at three a positions where charged residues are found instead of hydrophobic residues. In addition, no coiled-coil structure is predicted in the C-terminal domain. Another structural feature of the GeBP/GPL Leu-zipper is the distance between it and the DNA-binding domain. Although Leu-zipper motifs in HD-ZIP or bZIP TFs are immediately adjacent to the DNA-binding domains, the spacing ranges from 92 (GPL2) to 209 (GPL3) amino acids in the GeBP/GPL proteins. The formation of all combinations of homo- and heterodimers in the GeBP/GPL family means they potentially have a high combinatorial flexibility to regulate target genes, which are so far unidentified. Indeed, in plants, heterodimeric protein interactions mediated by the Leu-zipper motif increase the repertoire of potential DNA-protein interactions (Weltmeier et al., 2006).
This work allows the classification of GeBP/GPL as Leu-zipper proteins. Because homeodomain Leu-zipper proteins represent a subset of the large homeodomain family in plants, we suggest that GeBP/GPL proteins represent a novel form of DNA-binding Leu-zipper proteins within this family of 21 members.
Overlapping Expression and Functional Redundancy of GeBP/GPL Genes
According to their GUS expression patterns, the three most similar genes, GeBP, GPL1, and GPL2, are specifically expressed in aerial parts of the plant. There is a good correlation between the expression and function of these genes because cytokinin-related phenotypes were only visible in assays of aerial development, whereas none of the single, double, or triple mutants were affected in their root development or their cytokinin-sensitivity in root growth assays. Furthermore, the effects of gebp1, gpl1, and gpl2 mutations were generally additive in our experiments providing evidence for functional overlap within the family. One exception is the dark-induced leaf senescence assay where the triple mutant was not as distinguishable from the single and double mutants as in the other physiological or molecular assays. In this assay, however, leaves were separated from the main plant, and the SAM, where GeBP/GPL genes are mainly expressed, was not present. Therefore the functional redundancy of these genes in isolated leaves might be less striking. The expression of GeBP/GPL genes in leaf/cotyledon vascular tissues, hydathodes, and pedicel distal bulges suggests a role in vascular development. Cytokinins play an important role in the regulation of protoxylem formation in roots (Ye, 2002; Hutchison et al., 2006; Mahonen et al., 2006; Yokoyama et al., 2007), and many ARR genes are expressed in leaf vasculature (Ferreira and Kieber, 2005) or pedicel bulges, such as ARR5 (D'Agostino et al., 2000), in addition to their root expression.
Roles of GeBP/GPL Genes in Cytokinin Feedback Regulation
Type-A ARRs are considered to be primary cytokinin response genes that act as redundant negative regulators of cytokinin signaling and their transcription is rapidly elevated in response to exogenous cytokinin (D'Agostino et al., 2000; Kiba et al., 2002; Rashotte et al., 2003). Three lines of evidence indicate that the GeBP/GPL genes are involved in cytokinin responses through type-A ARR gene regulation. First, the triple mutant is less sensitive to exogenous cytokinins and has higher levels of type-A ARR transcripts. One important inference from these observations is that GeBP/GPL genes cannot belong to or be regulated by the phosphorelay cascade per se because the lower sensitivity of the triple mutant would then be coupled to a decrease in type-A ARR transcript levels as in cytokinin phosphorelay mutants (Hutchison et al., 2006). Second, transcript levels of type-A ARRs are partially insensitive to exogenous cytokinins in the triple mutant. This result suggests there is a nonadditive interaction between GeBP/GPL and cytokinin-dependent regulation of type-A ARRs. One hypothesis is that the GeBP/GPL genes interfere with the possible direct induction of type-A by type-B ARRs (Sakai et al., 2000; Rashotte et al., 2003). Third, 35S:VP16:GPL2 plants have reduced ARR transcript levels and display increased cytokinin sensitivity. This result strongly reinforces the involvement of GeBP/GPL genes in the promotion of cytokinin responses. One additional observation that supports the regulation of ARR genes by GeBP/GPL genes is the similar expression pattern of the four GeBP/GPL genes and type-A ARR genes such as ARR5 and ARR6 in the SAM and pedicels (To et al., 2004; Jasinski et al., 2005; Yanai et al., 2005). Taken together, these results strongly support the hypothesis that the function of GeBP/GPL genes is to regulate ARR expression and that this regulation occurs through at least one unknown repressor acting genetically downstream of GeBP/GPL. Type-A ARRs are the most highly cytokinin-responsive genes in Arabidopsis. Response genes other than type-A ARRs show weaker inductions, and data sets from different microarray experiments aimed at identifying transcriptional targets of cytokinin appear to vary considerably except for type-A ARRs (Muller and Sheen, 2007). In our conditions, transcript levels of such response genes (see “Materials and Methods”) were not significantly different in the triple mutant and the wild types. This is likely due to a weaker responsiveness of these genes and/or a modest modification of the cytokinin pathway in the triple mutant leading to even weaker changes in non-ARR response gene expression. Alternatively, we cannot exclude that only a subset of cytokinin response genes are modified in the triple mutant.
This function of GeBP/GPL genes in triggering the cytokinin response is in agreement with the role of cytokinin in SAM function. In Arabidopsis, cytokinin biosynthesis is necessary for SAM function and is positively regulated by KNOX genes (Jasinski et al., 2005). The KNOX gene BP has been shown to activate both ARR5 (Yanai et al., 2005) and GeBP expression (Curaba et al., 2003). Therefore, it might appear contradictory that BP increases ARR transcript levels while GeBP participates in their down-regulation. However, the induction of ARRs by BP is secondary to its effect on cytokinin accumulation. As mentioned above, this suggests that GeBP/GLP genes act on ARR expression through a cytokinin-independent mechanism. The simplest hypothesis is that the repression of ARRs by GeBP/GPL genes balances their induction by cytokinin thus enhancing cytokinin responses. From this point of view, GeBP/GPL genes act in a similar way to WUSCHEL (WUS), a homeodomain gene expressed in the central zone of the SAM, which also represses type-A ARRs (Leibfried et al., 2005) in the SAM. A major distinction is that WUS acts directly on ARR gene expression whereas GeBP/GPL genes seem to act through at least one additional, as yet unknown, repressor. This indirect regulation of ARRs raises the possibility that GeBP/GPL genes participate in other developmental pathways. Indeed, type-A ARRs are also regulated by the GA pathway (Greenboim-Wainberg et al., 2005) and they have been involved in light signal transduction (To et al., 2004) and in the control of circadian period (Salome et al., 2006). Therefore, we cannot exclude that GeBP/GPL genes play a role in the cross talk between these pathways. Also the change in type-A ARR expression could be a contributing factor to the phenotype but not the only factor.
Our analysis of the GeBP/GPL genes has led us to uncover their role in the regulation of the cytokinin response, namely the down-regulation of the negative feedback loop in cytokinin signaling. Future work is needed to determine whether GeBP family members that lack the Leu-zipper motif also participate in hormonal regulation in Arabidopsis and other plant species such as rice (Oryza sativa) and potato that have orthologous genes.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds were sown on soil or surface-sterilized and grown in petri dishes on MS Basal Salt Mixture medium (Sigma). Plants were grown at 22°C in long days (16 h of 100 μE light). The Arabidopsis (Arabidopsis thaliana) Landsberg erecta (Ler) and Columbia (Col-0) ecotypes were the wild types used. The gebp-1 line having a Ds transposon insertion in the GeBP locus (Ler background) has been described previously (Curaba et al., 2003). T-DNA insertion lines gpl1-1 (WiscDsLox391A04), gpl2-2 (SALK_054183), and gpl3-3 (SAIL_885_B10) were obtained from the Arabidopsis Biological Resource Center (Ohio State University) or the Nottingham Arabidopsis Stock Centre and are in the Col-0 background. Agrobacterium tumefaciens C58 pGV3121 was used for stable transformation of Arabidopsis (Col-0 ecotype) using the floral dip technique (Clough and Bent, 1998) or transient expression in tobacco leaves (Nicotiana benthamiana; Lavy et al., 2002).
Yeast Two-Hybrid and Immunoprecipitation Assays
The MatchMaker III system (CLONTECH) was used for yeast two-hybrid experiments. GeBP and GLP cDNAs were cloned from Gateway entry vectors into both pGADT7 and pGBKT7 vectors between EcoRI and XhoI sites and between EcoRI and SalI sites, respectively. Because yeast growth on permissive medium was impaired by strong expression of GeBP and GPL proteins, the long constitutive promoter PADH in the pGBKT7 and pGADT7 vectors was replaced by the short version from pGAD10 (CLONTECH). Yeast strain AH109 was cotransformed according to the manufacturer's instructions and selected on synthetic drop-out medium without Leu and Trp permissive medium. Individual colonies were grown in liquid culture and tested on synthetic drop-out medium supplement without Leu, Trp, adenine, and His selective medium supplemented with 5 to 10 mm 3-amino-1,2,4-triazole for 4 d at 30°C. The coiled-coil region in GeBP (Ala-176–Lys-209) was predicted with the program at http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_lupas.html (Lupas et al., 1991). Coimmunoprecipitation was done using the MatchMaker Co-IP kit (CLONTECH). Full-length cDNAs of GPL1 and GPL2 cloned into pGBKT7 and pGADT7 vectors, as described above, were used and proteins were synthesized and radiolabeled with the TnT T7 coupled reticulocyte lysate system (Promega). After immunoprecipitation, proteins were separated by SDS-PAGE and the gel was autoradiographed. Myc-tagged GPL2 and HA-tagged GPL1 were translated separately in vitro in the presence of [35S]Met and immunoprecipitated with the corresponding anti-tag antibody. Translation mixes were combined in a 1:1 ratio.
The megaprimer extension technique (Sambrook and Russell, 2001) was used for mutagenesis of the Leu-zipper motif using the AD-GeBP vector as template (Curaba et al., 2003). Mutagenized amplicons were cloned back into the vector using SpeI and XbaI sites and sequenced.
Confocal Microscopy
BiFC vectors were kindly provided by Dr. François Parcy (University Joseph Fourier). The GeBP cDNA was cloned upstream or downstream of both N-terminal and C-terminal fragments of the YFP gene using the Gateway cloning system (Invitrogen). Fusions were under the control of the 35S promoter. The four expression vectors were introduced separately into Agrobacterium and the four combinations of N-YFP and C-YFP fusions were independently coinfiltrated into tobacco leaves as previously described (Lavy et al., 2002) except that DAPI was added to the cell suspension at 1 μg mL−1 before infiltration. Observations were made with a Leica confocal microscope and data were analyzed with the Leica LCS 2.61 software. The N-YFP-GeBP and C-YFP-GeBP combination gave a specific YFP signal. Laser excitation was done in the sequential mode in between frames first with an argon laser (515 nm) and then with a UV laser (351–364 nm). Spectra were analyzed to confirm the specificity of YFP emission, which peaks around 527 nm.
Molecular Cloning
Oligonucleotides used for PCR amplification are given in Supplemental Table S1 online. GeBP/GPL cDNAs were produced from total RNA from 3-week-old rosettes. Genomic DNAs were cloned into pENTR/D-TOPO vector (Invitrogen), whereas cDNAs and promoters were cloned into pDONR221 (Invitrogen) using BP clonase (Invitrogen). A GeBP cDNA with no stop codon was also generated in pDONR221 for BiFC constructs where either the C-terminal or the N-terminal part of YFP was downstream of GeBP. GFP fusion lines for intracellular localization were made by cloning cDNAs into the pH7WGF2.0 vector (Plant Systems Biology, VIB-Ghent University) using LR clonase (Invitrogen) and stable transformation of the constructs into Arabidopsis. Transcriptional fusion lines with the GUS reporter gene were made by cloning GeBP/GPL promoters into the pKGWFS7 vector (Plant Systems Biology, VIB-Ghent University) and stable transformation of Arabidopsis with the constructs. For each construct, at least seven GUS-staining lines were studied. For GUS staining, plants were incubated 8 to 12 h with GUS substrate and destained as described (Gallagher, 1992). The 35S:GPL2 and 35S:VP16:GPL2 constructs were done using the Alligator2 and the Alligator1 vector, respectively (http://www.isv.cnrs-gif.fr/jg/alligator/vectors.html), kindly provided by François Parcy (Grenoble, France). GPL2 was used to make the VP16 fusion because the GPL2 protein is more readily detectable in transgenic plants. Indeed, GeBP, GPL1, and GPL3 proteins were hardly detectable in transgenic lines transformed with constructs aimed at overexpressing native or VP16 fusions forms. Expression of proteins in transgenic lines was assessed by western blot with an anti-HA antibody.
Isolation of gebp/gpl Mutants
Arabidopsis T-DNA insertion lines were screened by DNA sequence comparison as T-DNA or Ds transposon insertion site information was made available by Salk Institute Genomic Analysis (http://signal.salk.edu/index.html) and Cold Spring Harbor Laboratory (http://genetrap.cshl.org/). Gene-specific primers were used in combination with T-DNA or Ds transposon-specific primers to identify and confirm insertions by PCR (see Supplemental Table S1). These primer combinations and gene-specific primer combinations flanking the sites of insertions were used to distinguish heterozygous from homozygous plants. Only lines homozygous for T-DNA insertions were used in subsequent assays. Regarding the GPL2 gene, it should be noted that The Arabidopsis Information Resource (TAIR) annotation for At5g14280 is composed of four exons with the first three exons encoding a peptide homologous to GeBP and the fourth exon corresponding to a putative C-terminal extension. However, no ESTs or cDNAs have been described that cover the putative exon3-exon4 junction, and we could not detect transcripts overlapping this junction by reverse transcription (RT)-PCR. In addition, two nonoverlapping TAIR ESTs (137F3XP and RAFL17-19-N04) and data from the Massively Parallel Signature Sequencing technique (http://mpss.udel.edu/at/) indicate that exon4 is transcribed independently of the GPL-like ORF. Therefore, the GPL2 gene is likely to be restricted to the first three exons of the At5g14280 annotation, and T-DNA insertions in the putative exon4 were not considered as GPL2 mutant lines.
Double-insertion mutants were generated by crossing two single-insertion mutants. The gebp-1 allele was used to construct double mutants in which the GeBP function is impaired. To obtain the gebp-1 gpl1-1 gpl2-1 triple mutant, gebp-1 gpl2-1 double homozygous plants were crossed to plants homozygous for gebp-1 and heterozygous for gpl1-1. Plants homozygous for gebp-1 and gpl1-1 alleles and heterozygous for the gpl2-1 allele were identified in the F2 generation and selfed to produce F3 plants among which gebp-1 gpl1-1 gpl2-1 triple homozygous mutants were identified. F4 to F6 generations were used for the experiments described here.
Cytokinin Response Assays
For growth in the presence of cytokinin, surface-sterilized seeds were sown in petri dishes containing MS medium supplemented with kinetin (10 μg mL−1; Sigma) and plants were grown for 20 d at 22°C in a long day (16 h of 100 μmol s−1 cm−2 light). Chlorophyll content was measured after methanol extraction at 665 and 652 nm as described previously (Porra et al., 1989). Conditions for dark-induced senescence were as follows. Seeds were sown on soil, kept at 4°C for 3 to 4 d and plants were grown for 14 d. Leaves were detached and floated on water supplemented with BA (Sigma) at the indicated concentrations for 10 d in the dark. The chlorophyll content of three replicates of five leaves was measured at each concentration.
For the callus induction assay, root segments were excised from 10-d-old seedlings grown on MS medium and were incubated in the presence of 2,4-dichlorophenoxyac etic acid (30 ng mL−1) and various concentrations of kinetin for 24 d as previously described (Higuchi et al., 2004; To et al., 2004; Hutchison et al., 2006) except that MS medium was used instead of glucose minimal medium.
For the ARR transcript level assay, surface-sterilized seeds were sown in petri dishes containing MS medium and plants were grown for 14 d. Some petri dishes were sprayed with trans-zeatin (Sigma) as previously described (Kim et al., 2006) and rosettes were collected after 1 h. Total RNAs were extracted and RT was done as described (Curaba et al., 2003) using random hexamers and the moloney murine leukemia virus reverse transcriptase (Promega). PCRs contained 0.4 μg of reversed transcribed total RNA in 100-μL reactions using the ARR5, ARR6, ARR7, or Actin8 primers as described (Kim et al., 2006) using Taq polymerase (Bioline). The PCR cycle was 30 s at 94°C, 1 min at 60°C, and 1 min at 72°C. Aliquots of 25 μL were taken after cycles 28, 30, 32, and 34 to assess exponential amplification and 5-μL samples were electrophoresed on 1% agarose gels. Gels were stained with ethidium bromide, digitally scanned under UV light under exposing conditions that provided nonsaturating signals, and the integrated density of each band was measured with Image J software (http://rsb.info.nih.gov/ij/) and subtracted from the background below and above each band. Transcript levels of response genes other than type-A ARRs, such as an AP2-related gene (At3g16770; Brenner et al., 2005), a zinc finger gene (At4g26150; Brenner et al., 2005), senescence-associated protein gene (At1g53885; Rashotte et al., 2006), and the steroid sulfotransferase SST1 (At1g13420; To et al., 2004), genes were also measured using the same approach except SST1 for which real-time PCR was used.
Significant changes were assessed using the ANOVA (P < 0.05) test with the StatEL software (ad Science). Single mutants gpl1, gpl2, and gpl3 together with the double mutants gpl1 gpl2 and arr1 arr12 were tested with Col as the control group. Double mutants gebp gpl1 and gebp gpl2 together with the triple mutant gebp gpl1gpl2 were tested with Col and Ler as the control group and were considered significantly different if their mean value was above or under both control groups. The single mutant gebp was tested with Ler using the Mann and Whitney test.
Arabidopsis Genome Initiative locus identifiers for the GeBP/GPL genes are as follows: GeBP (At4g00270), GPL1 (At2g25650), GPL2 (At5g14280), and GPL3 (At2g36340).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genomic structures of the Arabidopsis GeBP/GPL genes and mutants.
Supplemental Table S1. List of primers used in this work.
Supplementary Material
Acknowledgments
We thank Dr. G. Eric Schaller (Dartmouth College) for the gift of the arr1 arr12 double mutant. We thank François Parcy (Université Joseph Fourier) for providing the BiFc and Alligator vectors, Jean-Pierre Alcaraz (Université Joseph Fourier) for his help in sequencing, and Cécile Cottet (Université Joseph Fourier) for her help in confocal analysis.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Daniel Perazza (daniel.perazza@ujf-grenoble.fr) and Gilles Vachon (gilles.vachon@ujf-grenoble.fr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Bornberg-Bauer E, Rivals E, Vingron M (1998) Computational approaches to identify leucine zippers. Nucleic Acids Res 26 2740–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Kollmer I, Burkle L, Schmulling T (2005) Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J 44 314–333 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42 567–585 [DOI] [PubMed] [Google Scholar]
- Chen H, Banerjee AK, Hannapel DJ (2004) The tandem complex of BEL and KNOX partners is required for transcriptional repression of GA20ox1. Plant J 38 276–284 [DOI] [PubMed] [Google Scholar]
- Chien JC, Sussex IM (1996) Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol 111 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Curaba J, Herzog M, Vachon G (2003) GeBP, the first member of a new gene family in Arabidopsis, encodes a nuclear protein with DNA-binding activity and is regulated by KNAT1. Plant J 33 305–317 [DOI] [PubMed] [Google Scholar]
- Curaba J, Moritz T, Blervaque R, Parcy F, Raz V, Herzog M, Vachon G (2004) AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol 136 3660–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino IB, Deruere J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira FJ, Kieber JJ (2005) Cytokinin signaling. Curr Opin Plant Biol 8 518–525 [DOI] [PubMed] [Google Scholar]
- Gallagher SR (1992) Quantitation of GUS activity by fluorometry. In SR Gallagher, ed, GUS Protocols Using the GUS Gene as a Reporter of Gene Expression. Academic Press, San Diego, pp 47–59
- Gan S, Amasino RM (1995) Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270 1986–1988 [DOI] [PubMed] [Google Scholar]
- Gan Y, Liu C, Yu H, Broun P (2007) Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 134 2073–2081 [DOI] [PubMed] [Google Scholar]
- Giulini A, Wang J, Jackson D (2004) Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430 1031–1034 [DOI] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D (2005) Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M (2002) The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr Biol 12 1557–1565 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mahonen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell SH, Lall S, Che P (2003) Cytokinins and shoot development. Trends Plant Sci 8 453–459 [DOI] [PubMed] [Google Scholar]
- Hutchison CE, Kieber JJ (2002) Cytokinin signaling in Arabidopsis. Plant Cell 14 S47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, et al (2006) The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413 383–389 [DOI] [PubMed] [Google Scholar]
- Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M (2005) KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15 1560–1565 [DOI] [PubMed] [Google Scholar]
- Kiba T, Yamada H, Mizuno T (2002) Characterization of the ARR15 and ARR16 response regulators with special reference to the cytokinin signaling pathway mediated by the AHK4 histidine kinase in roots of Arabidopsis thaliana. Plant Cell Physiol 43 1059–1066 [DOI] [PubMed] [Google Scholar]
- Kiba T, Yamada H, Sato S, Kato T, Tabata S, Yamashino T, Mizuno T (2003) The type-A response regulator, ARR15, acts as a negative regulator in the cytokinin-mediated signal transduction in Arabidopsis thaliana. Plant Cell Physiol 44 868–874 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I (2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA 103 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445 652–655 [DOI] [PubMed] [Google Scholar]
- Lavy M, Bracha-Drori K, Sternberg H, Yalovsky S (2002) A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14 2431–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438 1172–1175 [DOI] [PubMed] [Google Scholar]
- Long TA, Benfey PN (2006) Transcription factors and hormones: new insights into plant cell differentiation. Curr Opin Cell Biol 18 710–714 [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J (1991) Predicting coiled coils from protein sequences. Science 252 1162–1164 [DOI] [PubMed] [Google Scholar]
- Mahonen AP, Higuchi M, Tormakangas K, Miyawaki K, Pischke MS, Sussman MR, Helariutta Y, Kakimoto T (2006) Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr Biol 16 1116–1122 [DOI] [PubMed] [Google Scholar]
- Mason JM, Schmitz MA, Muller KM, Arndt KM (2006) Semirational design of Jun-Fos coiled coils with increased affinity: universal implications for leucine zipper prediction and design. Proc Natl Acad Sci USA 103 8989–8994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE (2005) Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller B, Sheen J (2007) Advances in cytokinin signaling. Science 318 68–69 [DOI] [PubMed] [Google Scholar]
- Ogas J, Cheng JC, Sung ZR, Somerville C (1997) Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277 91–94 [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD (1991) A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67 483–493 [DOI] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D (1998) A genetic framework for floral patterning. Nature 395 561–566 [DOI] [PubMed] [Google Scholar]
- Perazza D, Vachon G, Herzog M (1998) Gibberellins promote trichome formation by up-regulating GLABROUS1 in Arabidopsis. Plant Physiol 117 375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975 384–394 [Google Scholar]
- Rashotte AM, Carson SD, To JP, Kieber JJ (2003) Expression profiling of cytokinin action in Arabidopsis. Plant Physiol 132 1998–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Mason MG, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ (2006) A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc Natl Acad Sci USA 103 11081–11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Mandel T, Kuhlemeier C (2000) Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond A, Lang A (1957) Effect of kinetin on protein content and survival of detached xanthium leaves. Science 125 650–65113421662 [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290 2105–2110 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JA (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283 1541–1544 [DOI] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Oka A (2000) Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 24 703–711 [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294 1519–1521 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Kamiya N, Ueguchi-Tanaka M, Iwahori S, Matsuoka M (2001) KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev 15 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salome PA, To JP, Kieber JJ, McClung CR (2006) Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell 18 55–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell D (2001) Molecular Cloning, a Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Shani E, Yanai O, Ori N (2006) The role of hormones in shoot apical meristem function. Curr Opin Plant Biol 9 484–489 [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Kiba T, Sakakibara H, Ueguchi C, Mizuno T, Sugiyama T (1998) Expression of Arabidopsis response regulator homologs is induced by cytokinins and nitrate. FEBS Lett 429 259–262 [DOI] [PubMed] [Google Scholar]
- To JPC, Haberer G, Ferreira FJ, Deruere J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi C, Sato S, Kato T, Tabata S (2001) The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol 42 751–755 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40 428–438 [DOI] [PubMed] [Google Scholar]
- Weltmeier F, Ehlert A, Mayer CS, Dietrich K, Wang X, Schutze K, Alonso R, Harter K, Vicente-Carbajosa J, Droge-Laser W (2006) Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J 25 3133–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O, Shani E, Dolezal K, Tarkowski P, Sablowski R, Sandberg G, Samach A, Ori N (2005) Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr Biol 15 1566–1571 [DOI] [PubMed] [Google Scholar]
- Ye ZH (2002) Vascular tissue differentiation and pattern formation in plants. Annu Rev Plant Biol 53 183–202 [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Yamashino T, Amano Y, Tajima Y, Imamura A, Sakakibara H, Mizuno T (2007) Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol 48 84–96 [DOI] [PubMed] [Google Scholar]
- Zourelidou M, de Torres-Zabala M, Smith C, Bevan MW (2002) Storekeeper defines a new class of plant-specific DNA-binding proteins and is a putative regulator of patatin expression. Plant J 30 489–497 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.