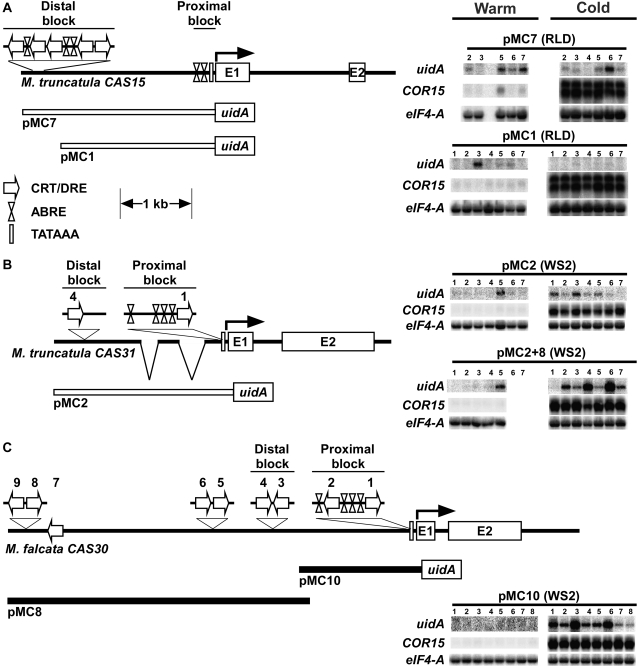
Figure 5.
RNA blot analysis of Medicago CAS promoter uidA fusions in Arabidopsis. A, MtCAS15 and the MtCAS15 promoter-uidA reporter gene fusion constructs that either included (pMC7), or excluded (pMC1) the distal upstream CRT/DRE and ABRE motifs (B) MtCAS31 and the MtCAS31 promoter-uidA reporter gene fusion constructs pMC2 and pMC2+8. C, MfCAS30 and the MfCAS30 promoter-uidA reporter gene fusion construct pMC10. Line drawings are drawn to scale, except for the clustered CRT/DRE and ABRE sequences, which are shown above the scaled horizontal genomic regions. CRT/DRE motifs in the MfCAS30 upstream region are numbered beginning with number 1 adjacent to the CDS. The numbering of CRT/DRE motifs in MtCAS31 is based on homology with MfCAS30 and follows that of the MfCAS30 scheme. The two breaks in MtCAS31 represent deletions relative to MfCAS30. Expression analysis conducted using T2 plants from multiple independent lines of each construct transformed into Arabidopsis laboratory strain RLD (pMC7 and pMC1) or WS-2 (pMC2, pMC10, and pMC2+8) and assayed for uidA, COR15, and eIF4-A transcript accumulation under noninducing conditions (warm) or after 6 h of low-temperature exposure (cold). Each lane contains 7 μg total RNA.