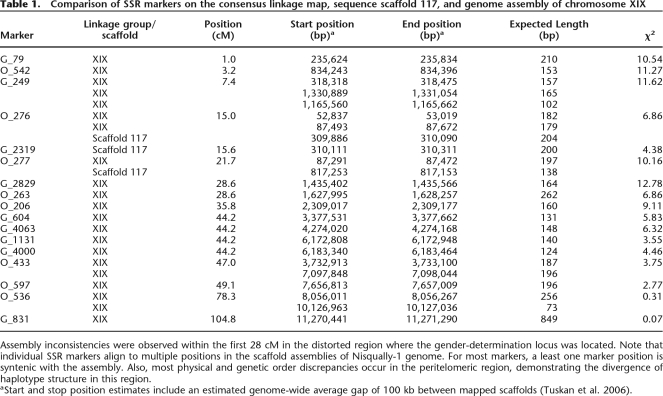
Table 1.
Comparison of SSR markers on the consensus linkage map, sequence scaffold 117, and genome assembly of chromosome XIX
Assembly inconsistencies were observed within the first 28 cM in the distorted region where the gender-determination locus was located. Note that individual SSR markers align to multiple positions in the scaffold assemblies of Nisqually-1 genome. For most markers, a least one marker position is syntenic with the assembly. Also, most physical and genetic order discrepancies occur in the peritelomeric region, demonstrating the divergence of haplotype structure in this region.
aStart and stop position estimates include an estimated genome-wide average gap of 100 kb between mapped scaffolds (Tuskan et al. 2006).