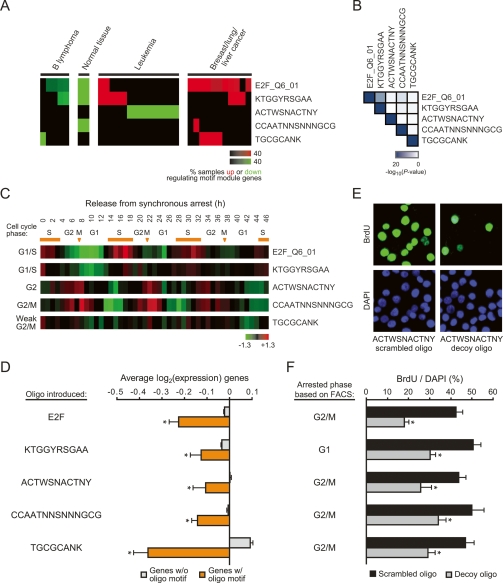
Figure 5.
Uncharacterized motifs regulate cell cycle progression. (A) Clinical annotation enrichment of the array signature of E2F_Q6_01 and the four uncharacterized motif modules. (B) Significance of enrichment of the indicated motif module target genes with all other motif targets is shown. (C) Shown is the average expression of the indicated motif module genes found in a set of periodically expressed cell cycle genes in HeLa cells (Whitfield et al. 2002). The stage of cell cycle that the pattern of expression most resembles is indicated on the left. Orange bars signify S phase; arrows signify mitoses. (D) Decoy oligos corresponding to the uncharacterized motif sequences inhibit target gene expression in HeLa cells. Shown is the average log2 gene expression (± SE) of genes predicted to have the indicated motif (orange) or those that lack the motif (gray). (*) P < 0.05 compared with genes without motif, Student’s t-test. (E,F) Decoy oligos inhibit cell cycle progression in HeLa cells. (E) Example of BrdU immunofluorescence staining and DAPI counterstain after introduction of scrambled oligos or decoy oligos for the ACTWSNACTNY motif. (F) Average percentage (± SE) of BrdU-positive cells after transfection of the indicated scrambled or decoy oligos. The cell cycle phase in which the cells arrested (as determined by FACS) is indicated on the left. (*) P < 0.01 compared with the respective scrambled oligo, Student’s t-test.